scales::label_percent()(c(0.4, 0.6, .95))[1] "40%" "60%" "95%"scales::label_comma()(20^(1:4))[1] "20" "400" "8,000" "160,000"scales::breaks_log()(20^(1:3))[1] 10 30 100 300 1000 3000 10000ggplot, part 2
MADS6
2026-02-25
Mapping is the function for associating a data column with an aesthetic.
Scales are the functions that associate a data value with a visual point.
Functions are based on this scheme:
scale_{aes}_{type}()Arguments to change default
breaks, choose where are labelslabels, usually with package scalestrans, to change to i.e log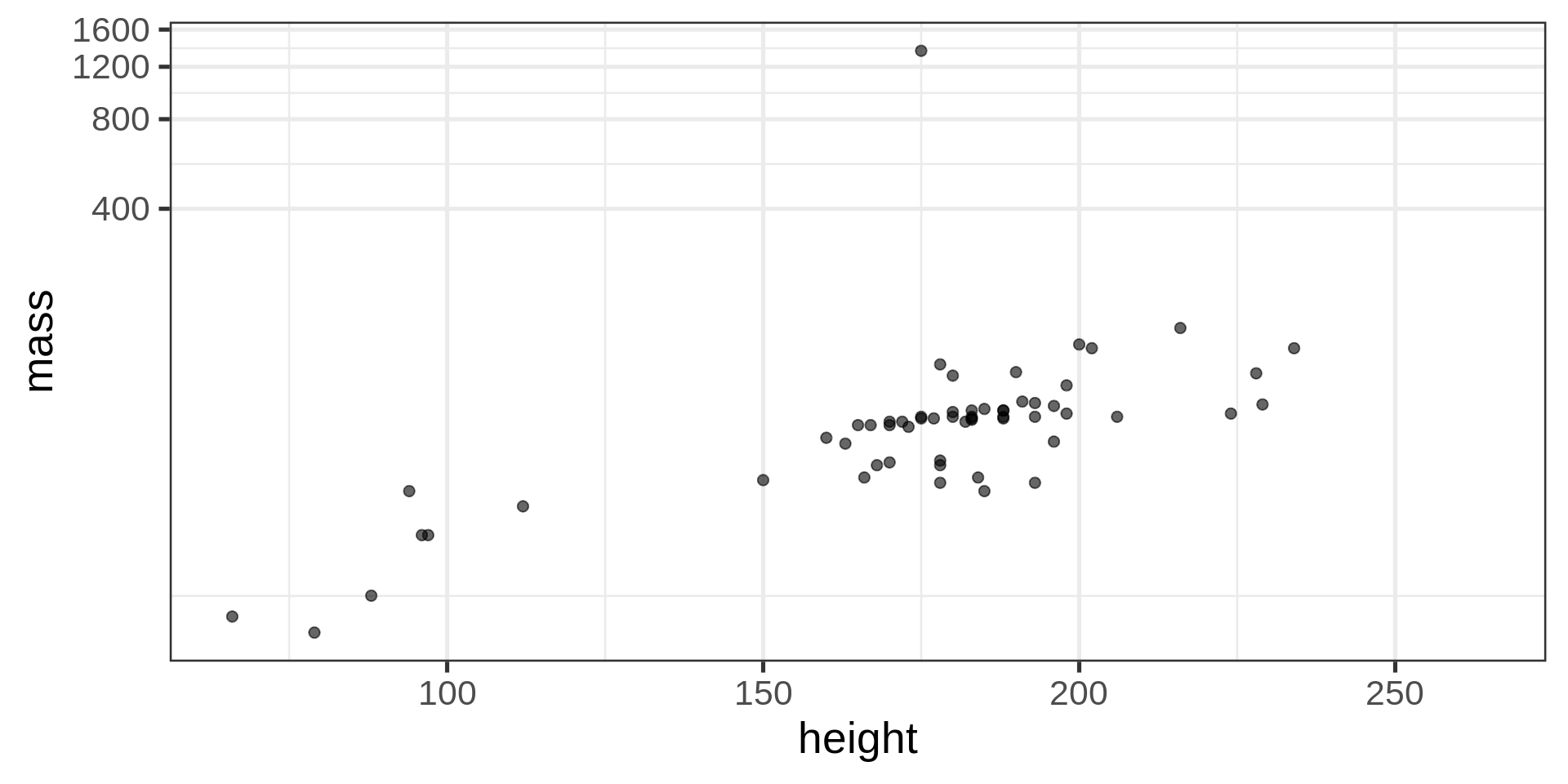
Tip
All geometries geom_ can take either a data.frame or a function, here geom_text().
Better: see Emil Hvitfeldt repo
scale_fill_hue()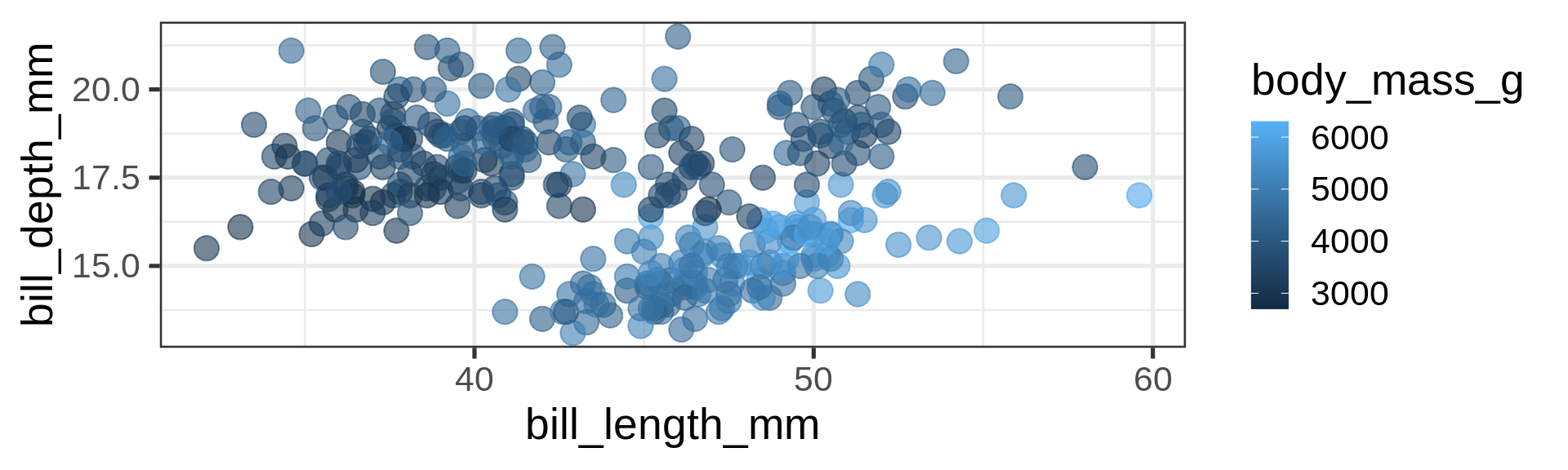
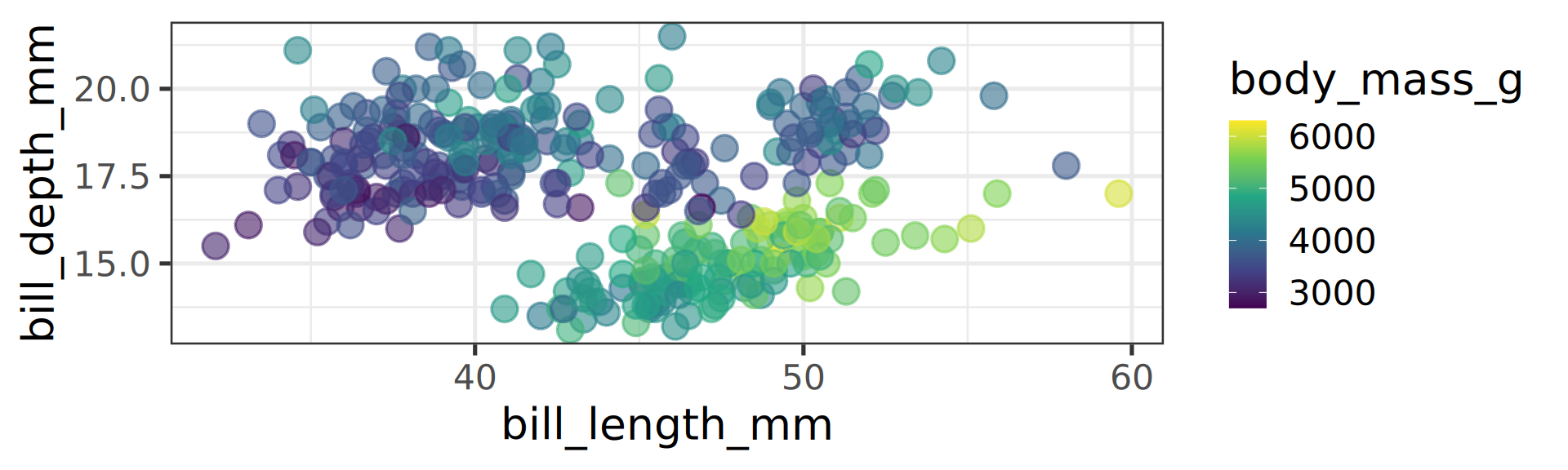
Viridis gradient is usually an improvement
Default ggplot2 is prints(!) well but on screens better off using viridis.
scale_color_viridis_c() c for continuous.

ggplot2 since v3.0 but no the default How to set the title
labs()labs()facet_wrap()facet_grid() to lay out panels in a grid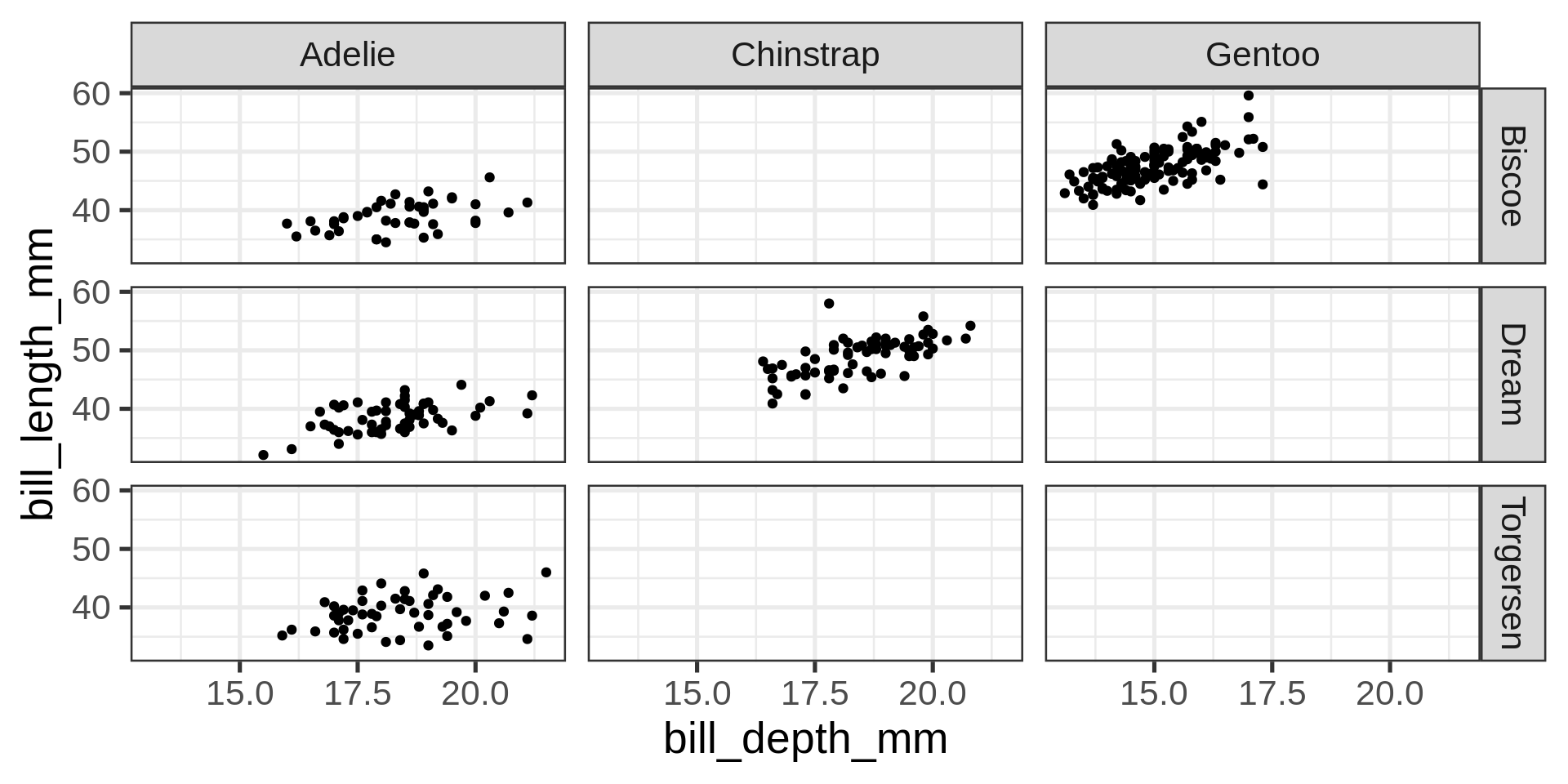
Grid lay out
~facet_grid() can also be used with one variable, complemented by a placeholder: .
ggplot2
Finding working extensions
ggplot2 introduced the possibility for the community to create extensions, they are referenced on a dedicated site
(https://exts.ggplot2.tidyverse.org/gallery/)
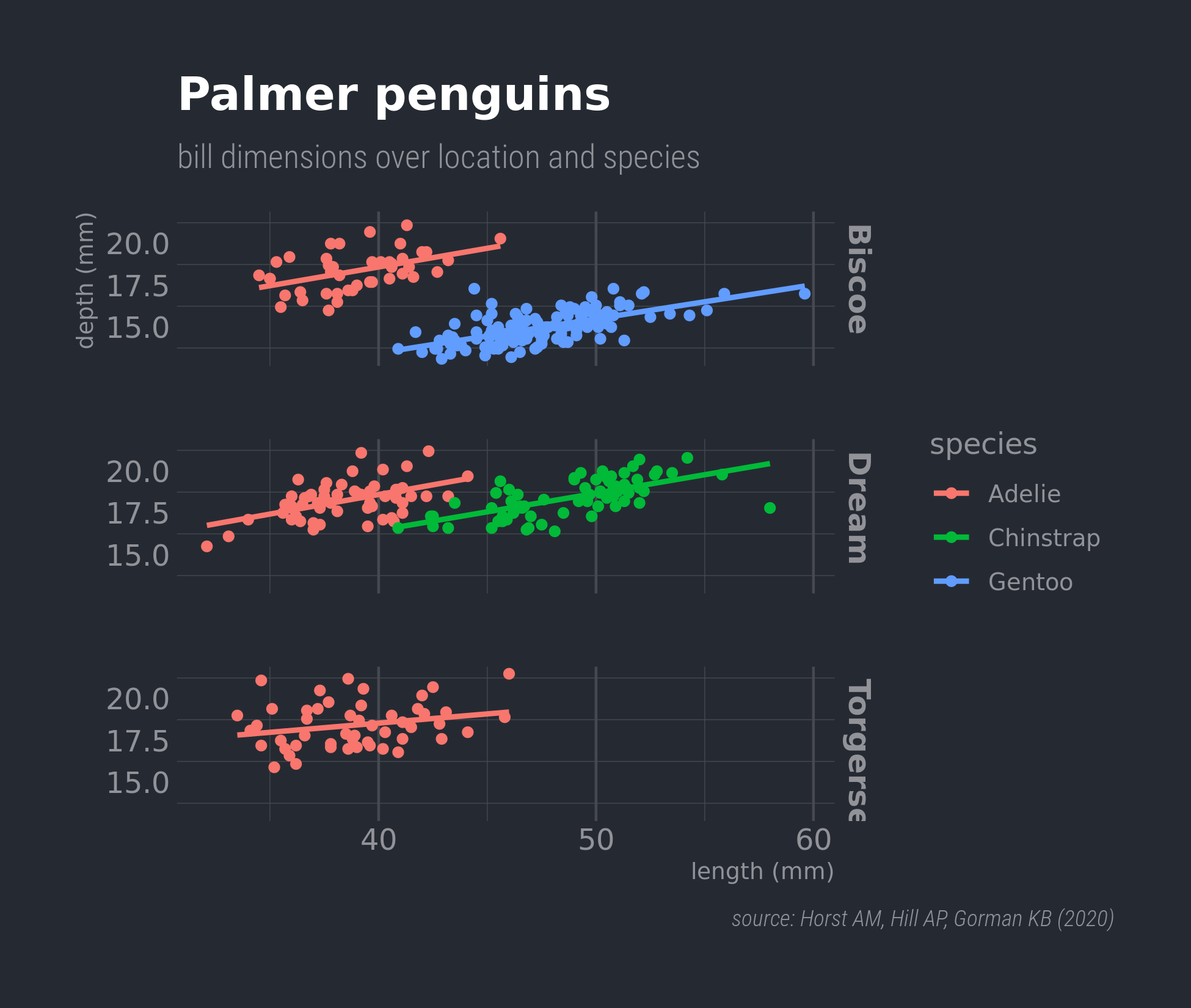
ggplot(penguins,
aes(x = bill_length_mm,
y = bill_depth_mm,
colour = species)) +
geom_point() +
geom_smooth(method = "lm", formula = 'y ~ x',
# no standard error ribbon
se = FALSE) +
facet_grid(island ~ .) +
labs(x = "length (mm)", y = "depth (mm)",
title = "Palmer penguins",
subtitle = "bill dimensions over location and species",
caption = "source: Horst AM, Hill AP, Gorman KB (2020)") +
theme_dark(14) +
# tweak the theme
theme(panel.grid.major.y = element_blank(),
panel.grid.major.x = element_line(size = 0.5),
panel.background = element_rect(fill = "grey10"),
plot.caption = element_text(face = "italic"),
strip.text = element_text(face = "bold"),
plot.caption.position = "plot")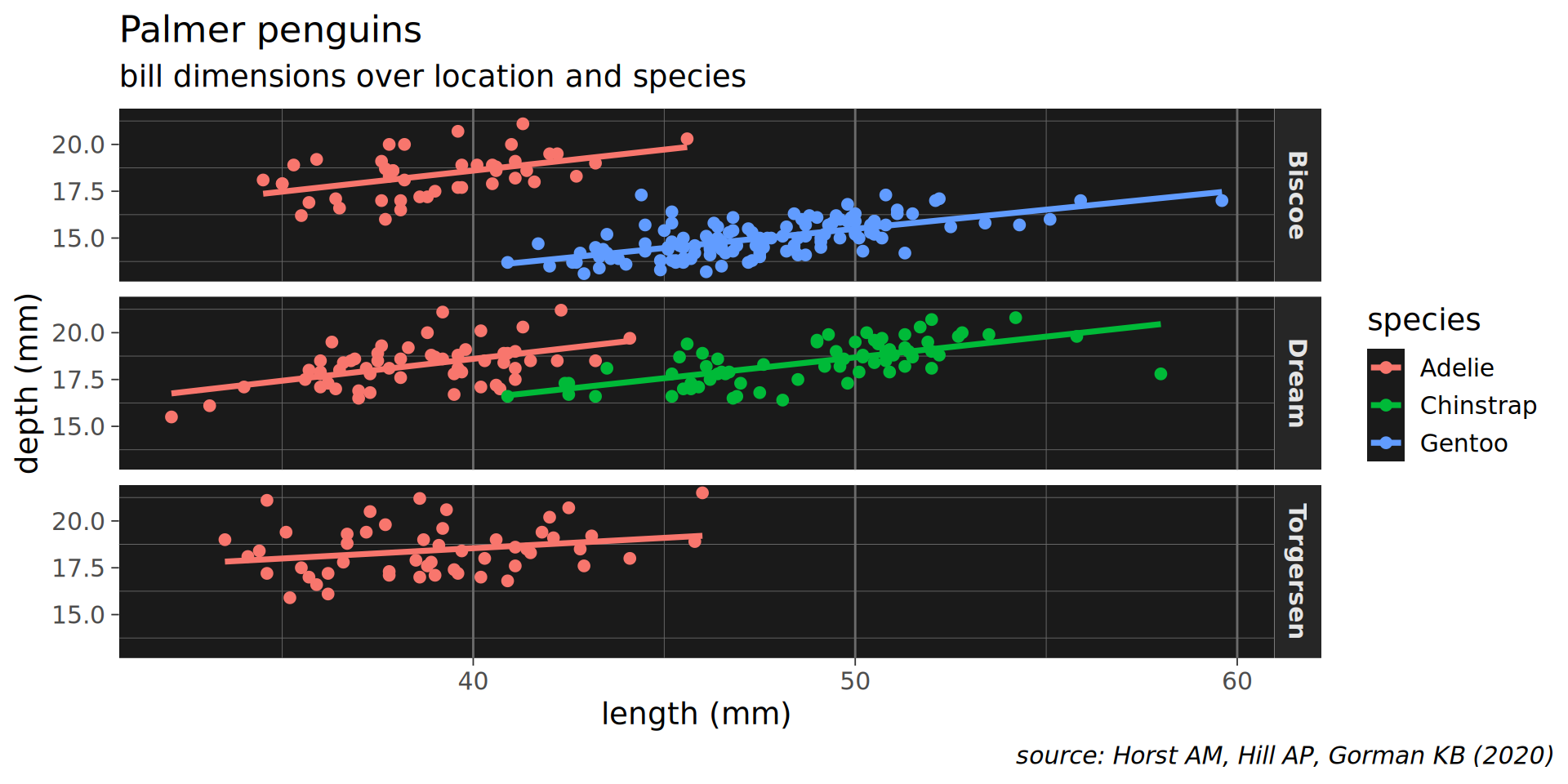
patchwork
Patchwork is developed by Thomas Lin Pedersen, maintainer of ggplot2.
patchwork
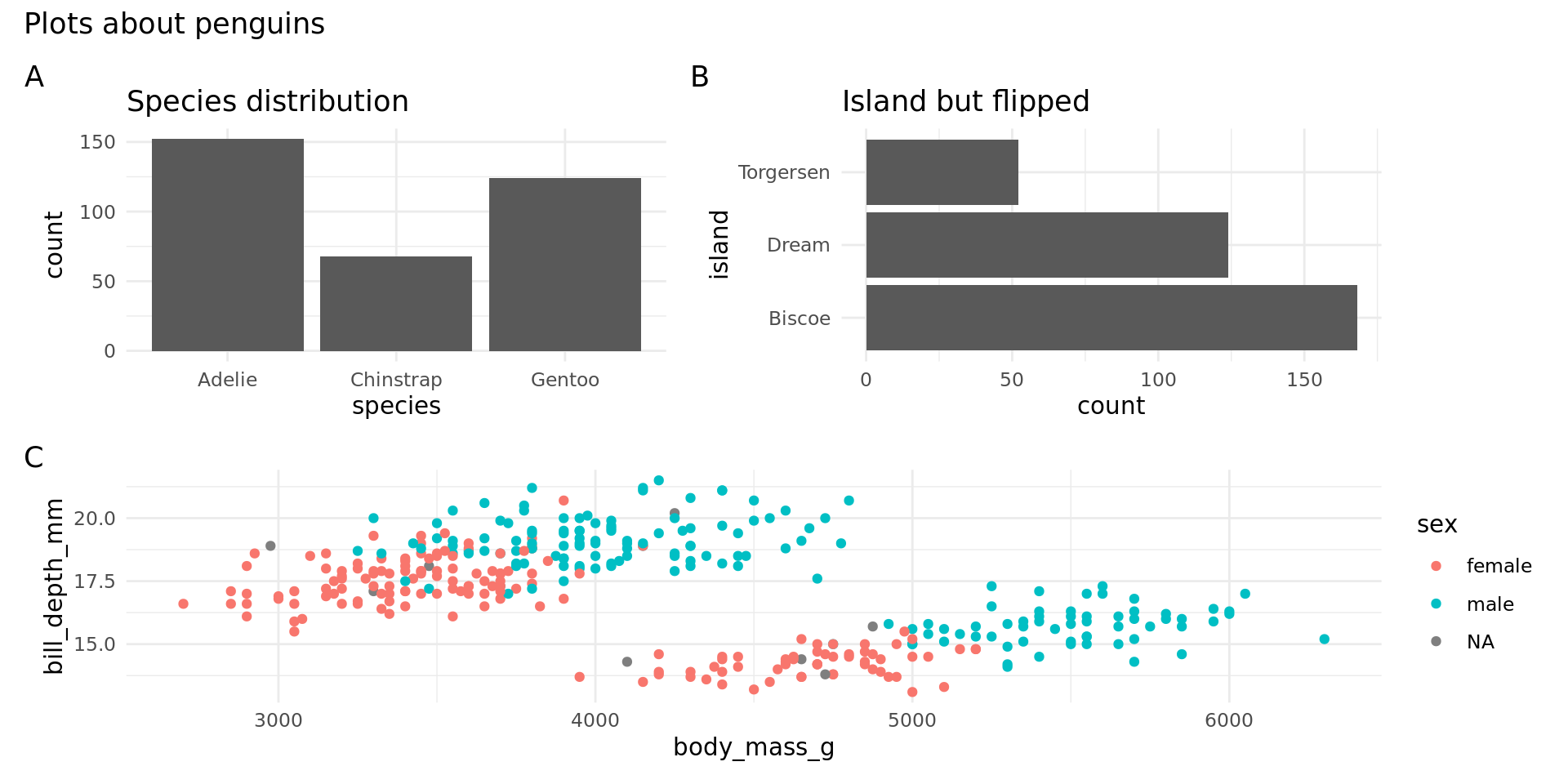
Composition
patchwork provides an API using the classic arithmetic operators
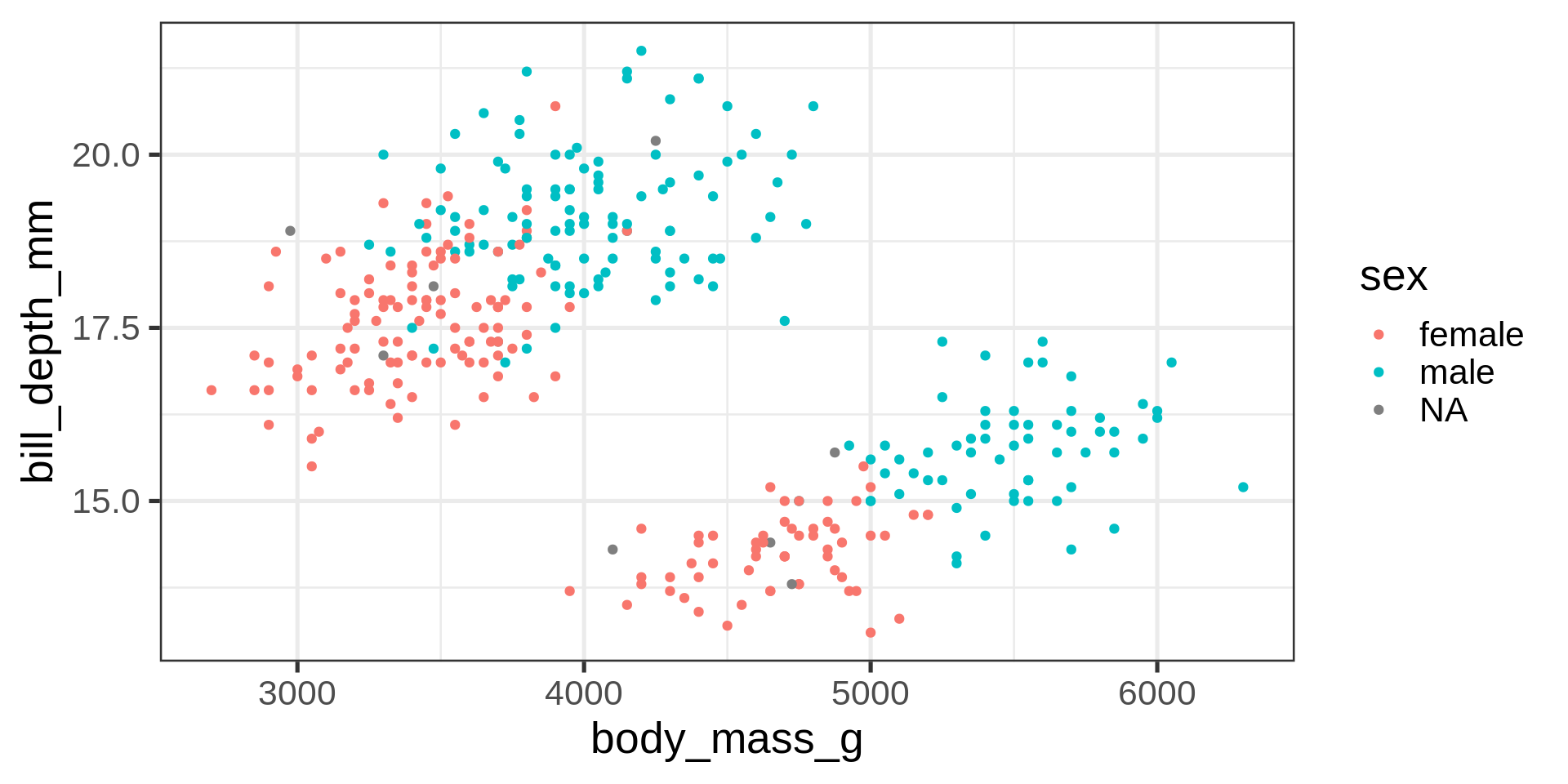
If you need to perfectly align axes, have a look at cowplot by Claus O. Wilke

Allan Cameron: geomtextpath package
ggplot object, 2nd argument, here p or last_plot()svg, png, svg, etc.)fig-height, fig-widthfig-asp…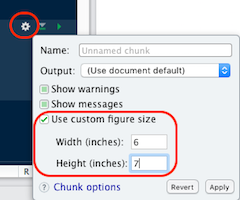
geom_tile() heatmapgeom_bind2d() 2D binninggeom_abline() slopestat_ellipse() (ggforge seen)stat_summary() easy mean, 95CI etc.ggforce::facet_grid_paginate() facetsgridExtra::marrangeGrob() plotscoord_cartesian() for zooming inesquisse Rstudio addin by dreamRs
You learned to:
Acknowledgments
*Further reading
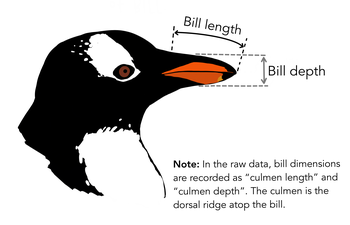
*: Palmer penguins, data are available by CC-0 license and Artwork by Allison Horst