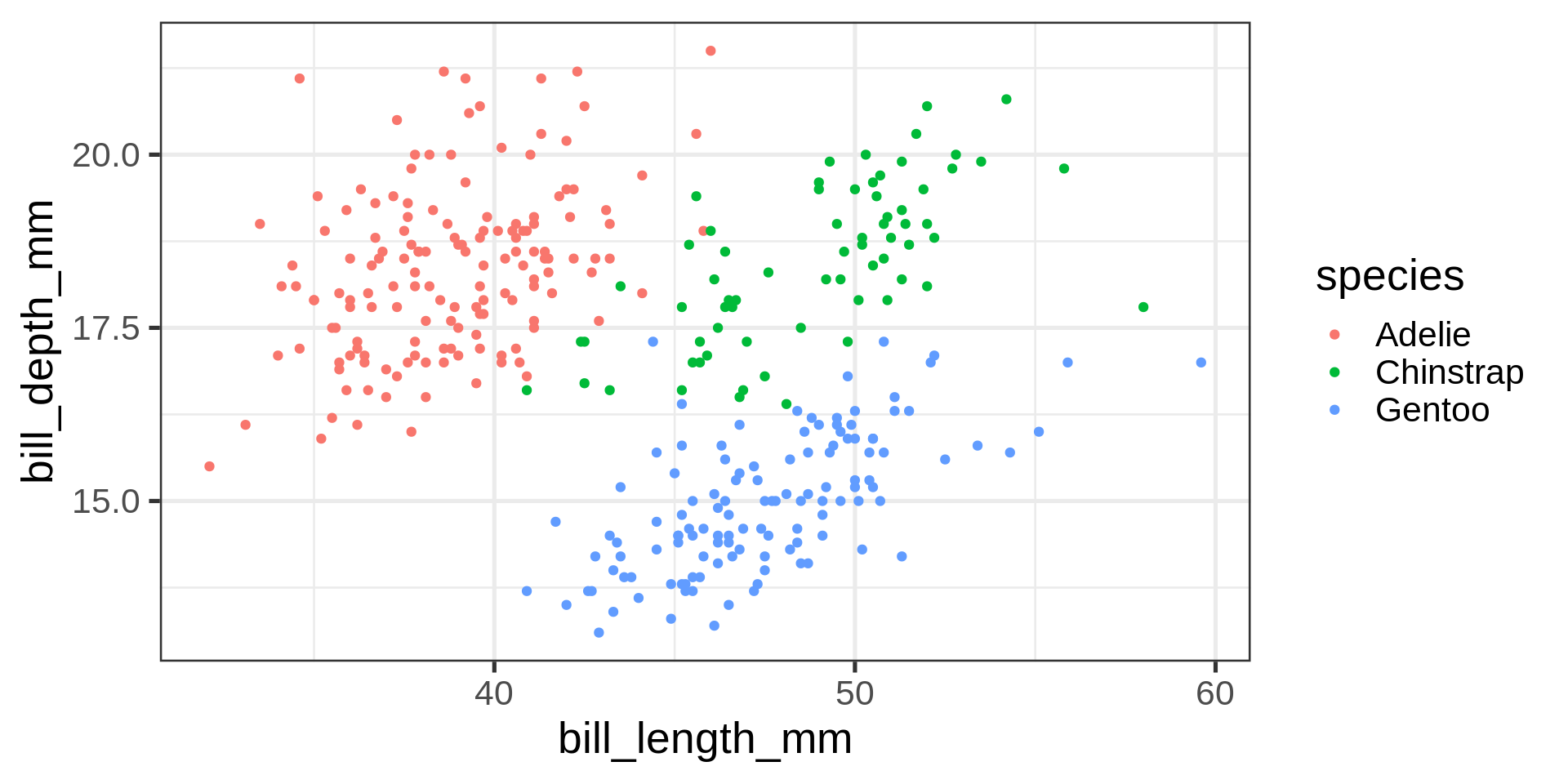
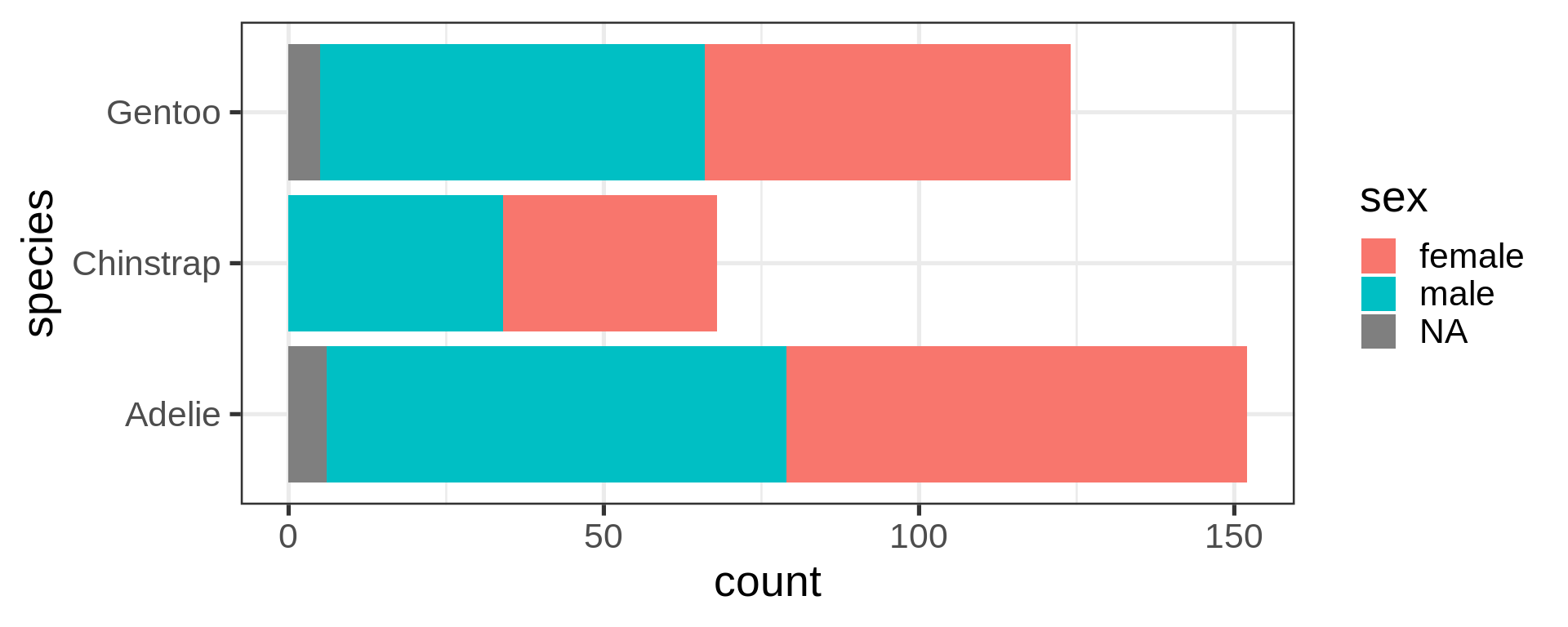
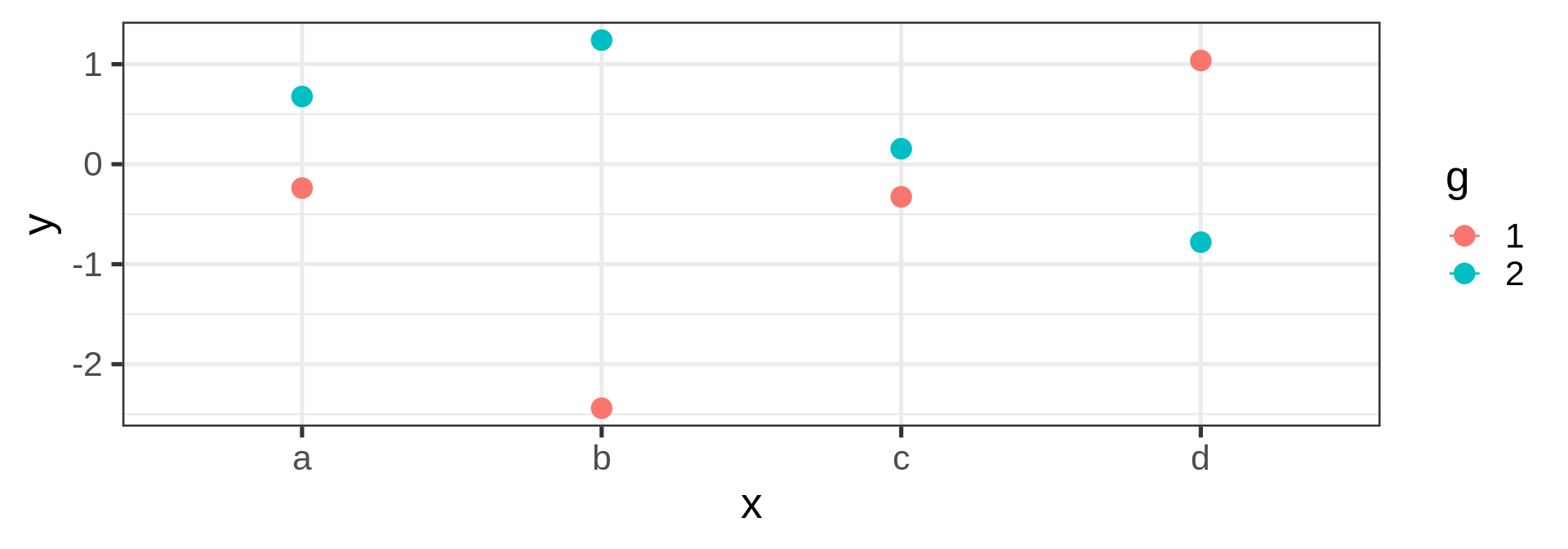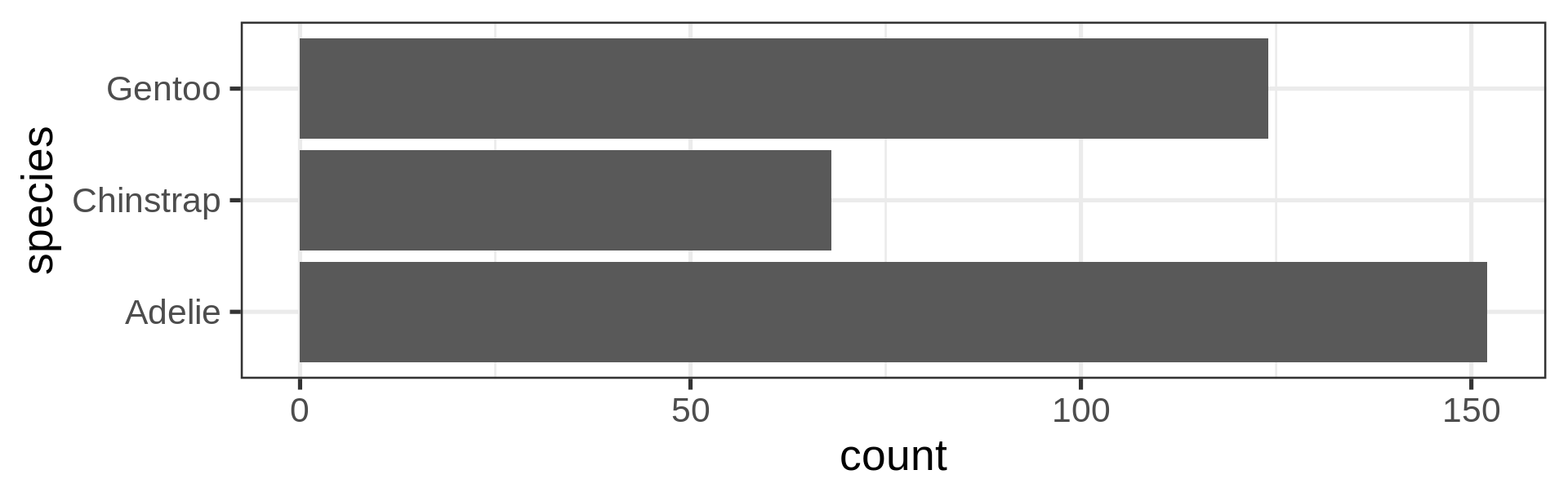library(ggplot2)
swiss |>
ggplot() +
aes(x = Education,
y = Examination) +
geom_point() +
scale_colour_brewer()Plotting data
ggplot, part 1
MADS6
2026-02-25
Introduction
Learning objectives
- Learn and apply the basic grammar of graphics
- Understand how it is implemented in
ggplot2 - Input data structures as
data.frame/tibble - Mapping columns to display features (aesthetics)
- Types of graphics (geometries)
Followed by a second lecture
Introduction
ggplot2
- Stands for grammar of graphics plot v2
- Inspired by Leland Wilkinson work on the grammar of graphics in 2005.
Graphs are split into layers
- Separate axis, curve(s), labels.
- 3 elements are required:
- data,
- aesthetics,
- geometry \(\geqslant 1\)
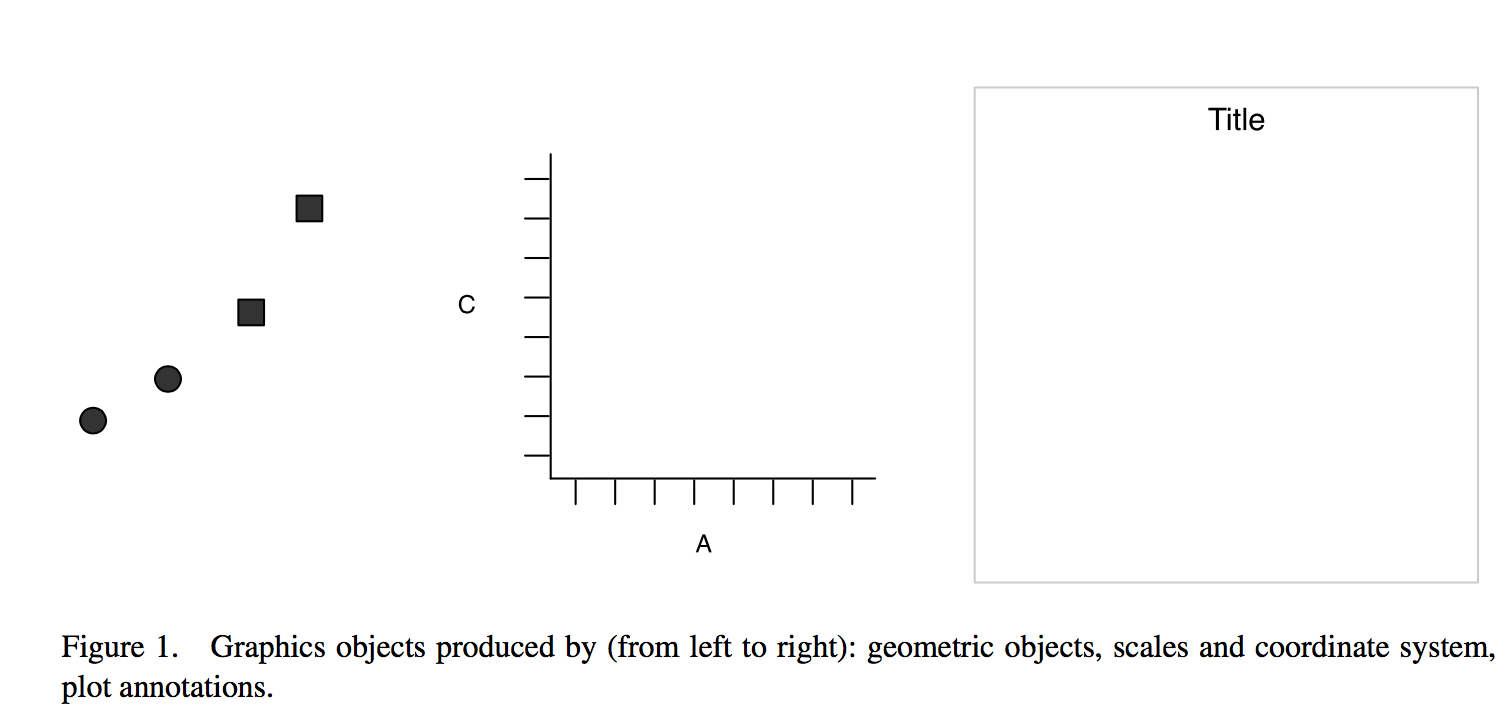
Creating a plot
Data
| A | B | C | D |
|---|---|---|---|
| 2 | 3 | 4 | a |
| 1 | 2 | 1 | a |
| 4 | 5 | 15 | b |
| 9 | 10 | 80 | b |
Aesthetics function
x = A
y = C
shape = D
Scaling to physical units \(x = \frac{A-min(A)}{range(A)} \times width\)
\(y = \frac{C-min(C)}{range(C)} \times height\)
\(shape = f_{s}(D)\)
Geometry
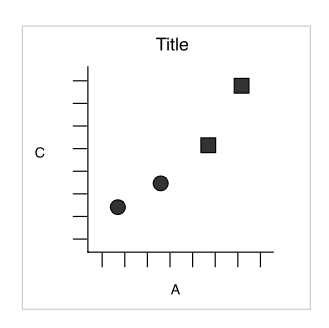
Scatter plot
Data drawn as points
Mapped data
| x | y | shape |
|---|---|---|
| 25 | 11 | circle |
| 0 | 0 | circle |
| 75 | 53 | square |
| 200 | 300 | square |
Wickham: A Layered Grammar of Graphics (2007)
Result
What if we want to split into panels circles and squares?
Faceting
Split by shape, aka trellis or lattice plots
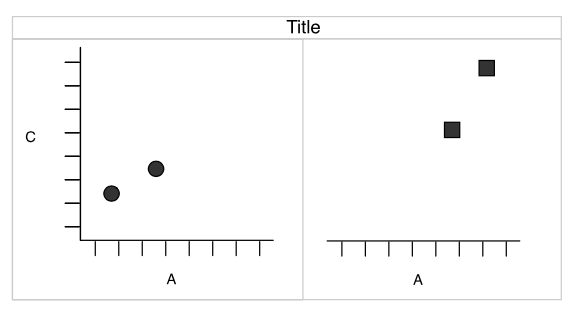
Redundancy
- shape and facets provide the same information.
- The
shapeaesthetic is free for another variable.
Wickham: A Layered Grammar of Graphics (2007)
Familiar country shape and data

Florence Débarre tweet, 2021-12-19 about Omicron incoming wave. Of note, done with R base!
Combining layers

All ggplot layers are functions
Warning
ggplot2layers are combined with+!- The magrittr pipe
%>%or the base pipe|>will not work! - This introduces a break in the workflow.
Palmer penguins

 Horst AM, Hill AP, Gorman KB (2020). palmerpenguins: Palmer Archipelago (Antarctica) penguin data. R package
Horst AM, Hill AP, Gorman KB (2020). palmerpenguins: Palmer Archipelago (Antarctica) penguin data. R package v0.1.0
Capabilities in ggplot
# A tibble: 344 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
7 Adelie Torgersen 38.9 17.8 181 3625
8 Adelie Torgersen 39.2 19.6 195 4675
9 Adelie Torgersen 34.1 18.1 193 3475
10 Adelie Torgersen 42 20.2 190 4250
# ℹ 334 more rows
# ℹ 2 more variables: sex <fct>, year <int>Capabilities in ggplot
Capabilities in ggplot
Capabilities in ggplot
Capabilities in ggplot
Capabilities in ggplot
Capabilities in ggplot
Capabilities in ggplot
Capabilities in ggplot
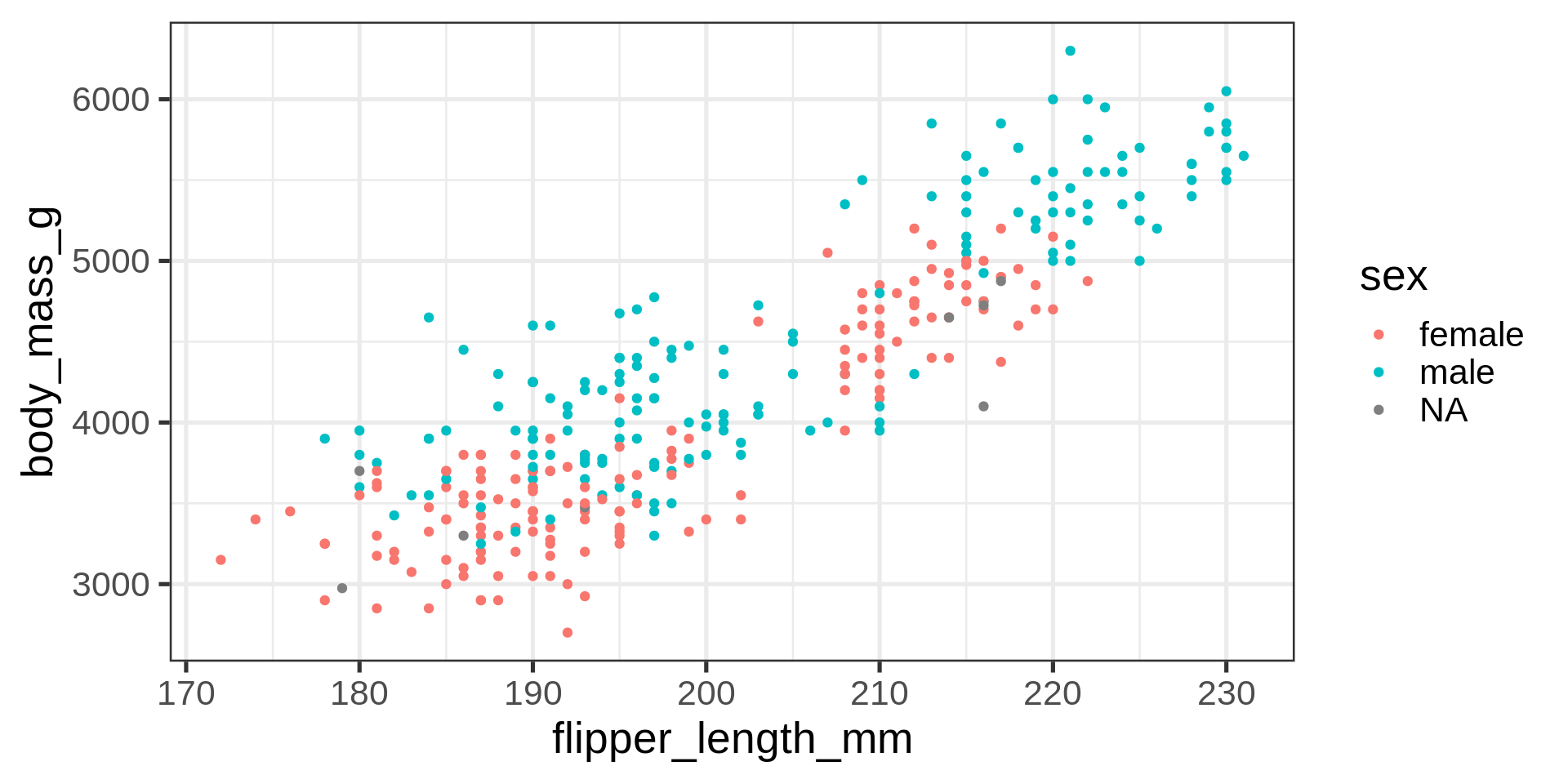
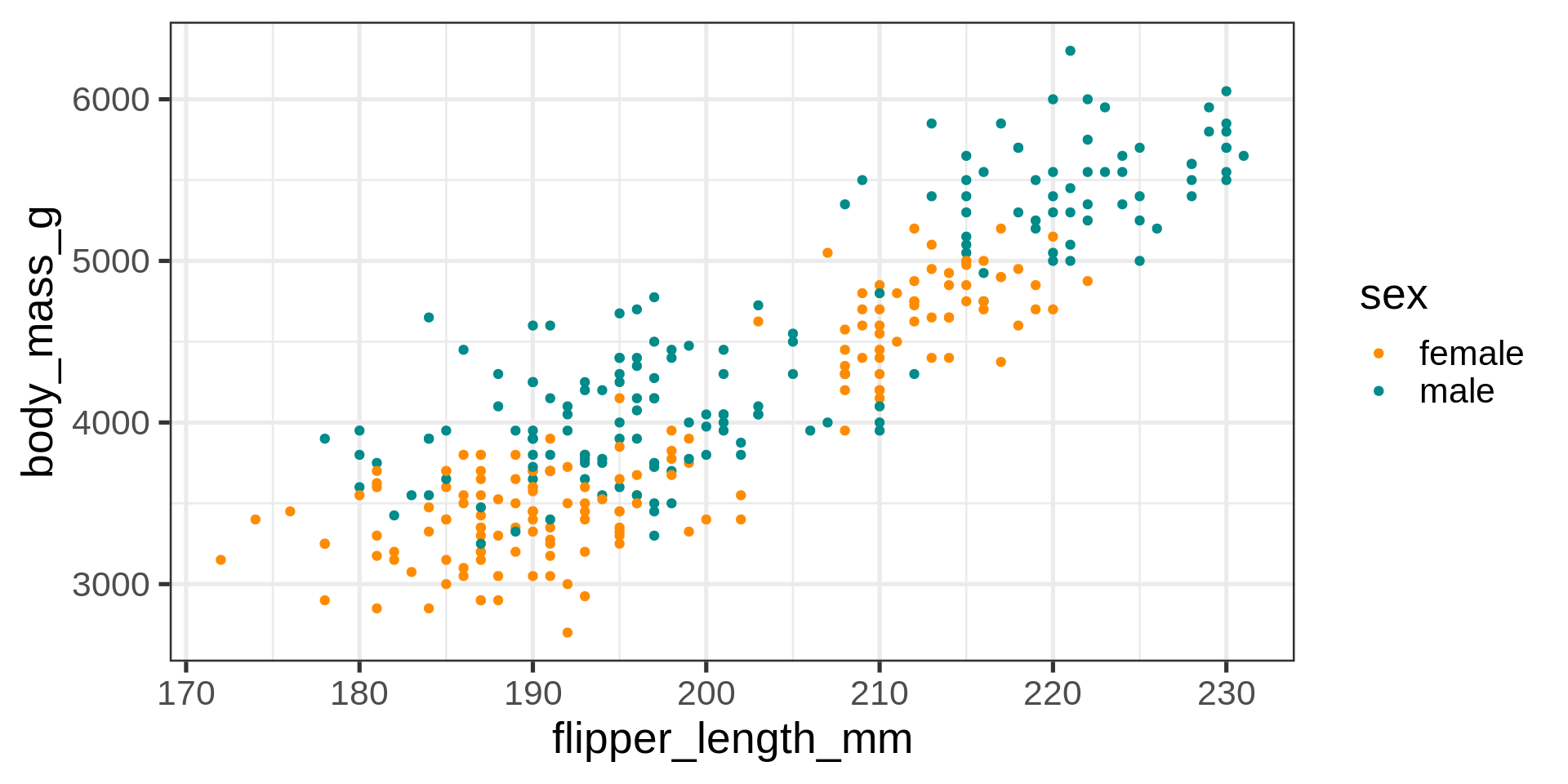
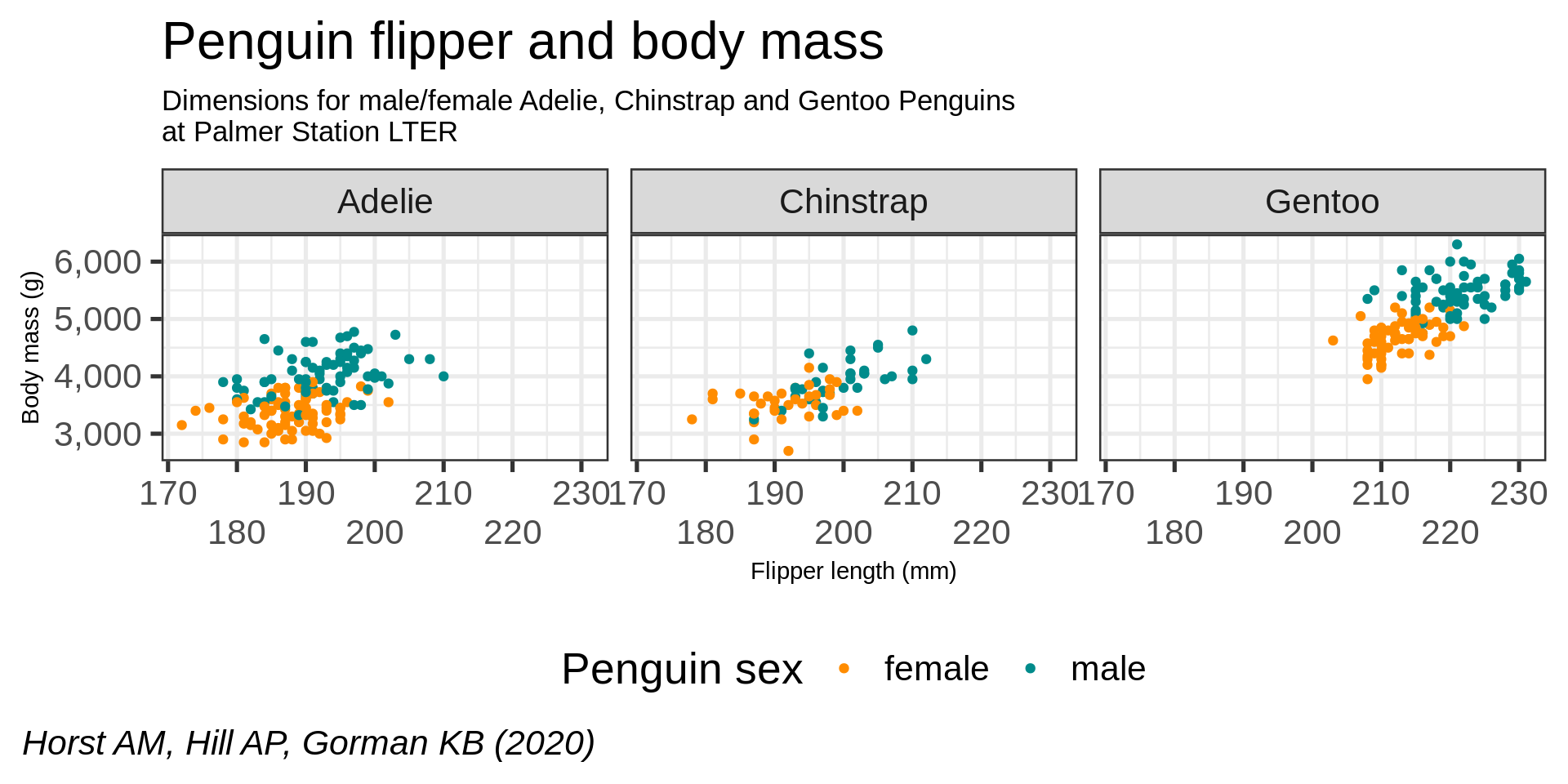
ggplot(data = penguins) +
aes(x = flipper_length_mm,
y = body_mass_g,
color = sex) +
geom_point() +
scale_color_manual(values = c("darkorange", "cyan4"),
na.translate = FALSE) +
labs(title = "Penguin flipper and body mass",
caption = "Horst AM, Hill AP, Gorman KB (2020)",
subtitle = description,
x = "Flipper length (mm)",
y = "Body mass (g)")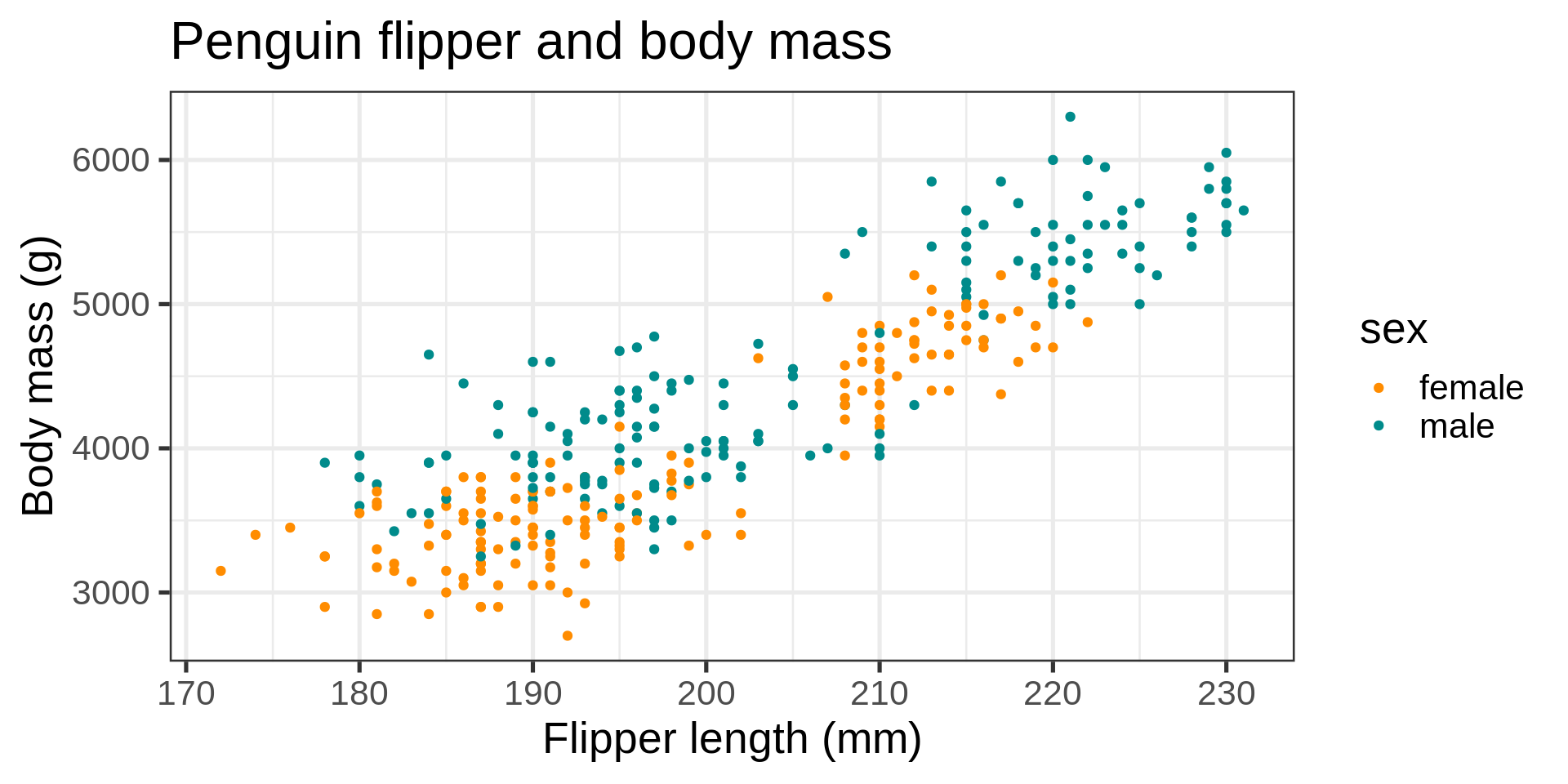
Capabilities in ggplot
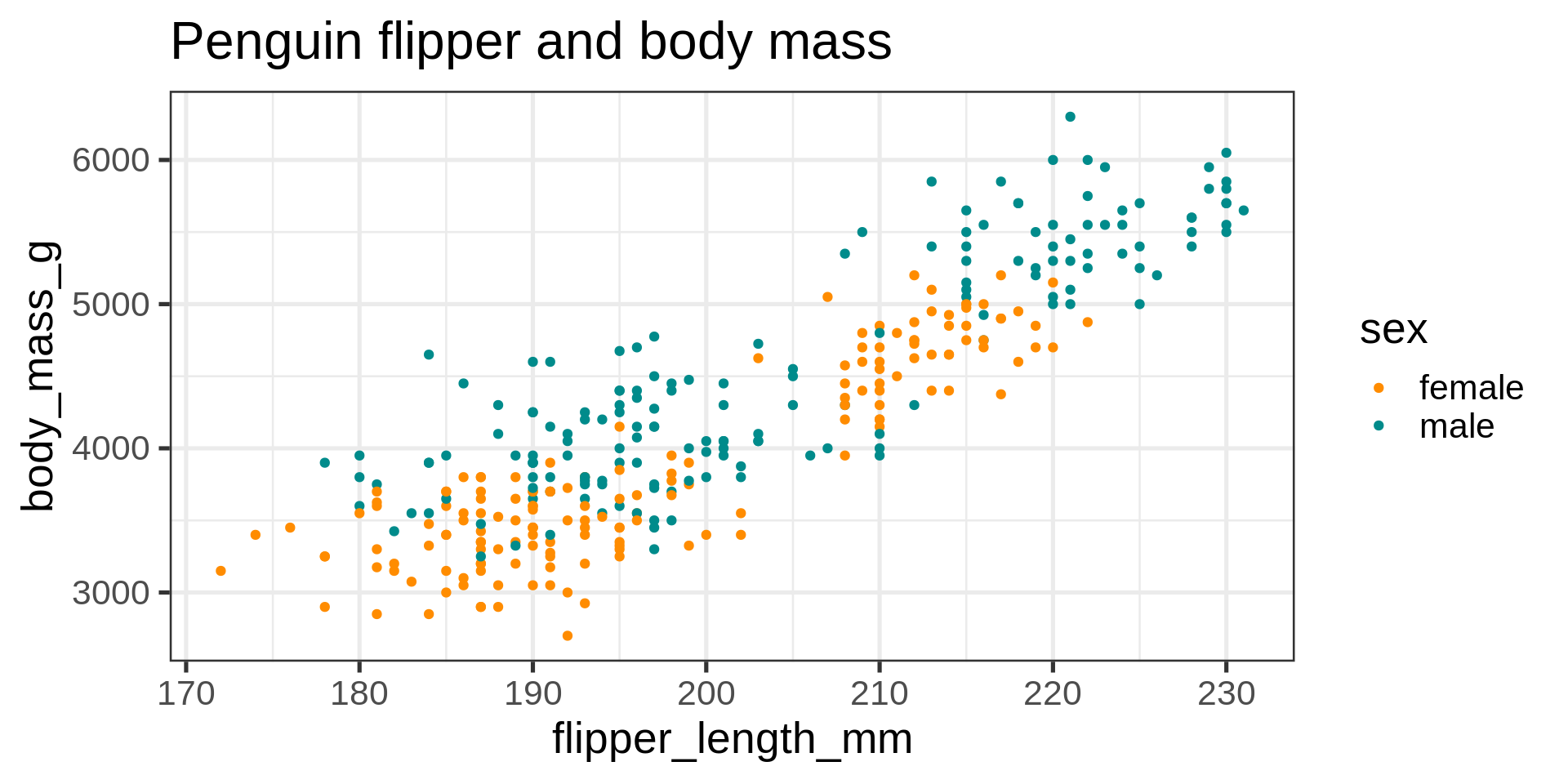
ggplot(data = penguins) +
aes(x = flipper_length_mm,
y = body_mass_g,
color = sex) +
geom_point() +
scale_color_manual(values = c("darkorange", "cyan4"),
na.translate = FALSE) +
labs(title = "Penguin flipper and body mass",
caption = "Horst AM, Hill AP, Gorman KB (2020)",
subtitle = description,
x = "Flipper length (mm)",
y = "Body mass (g)",
color = "Penguin sex")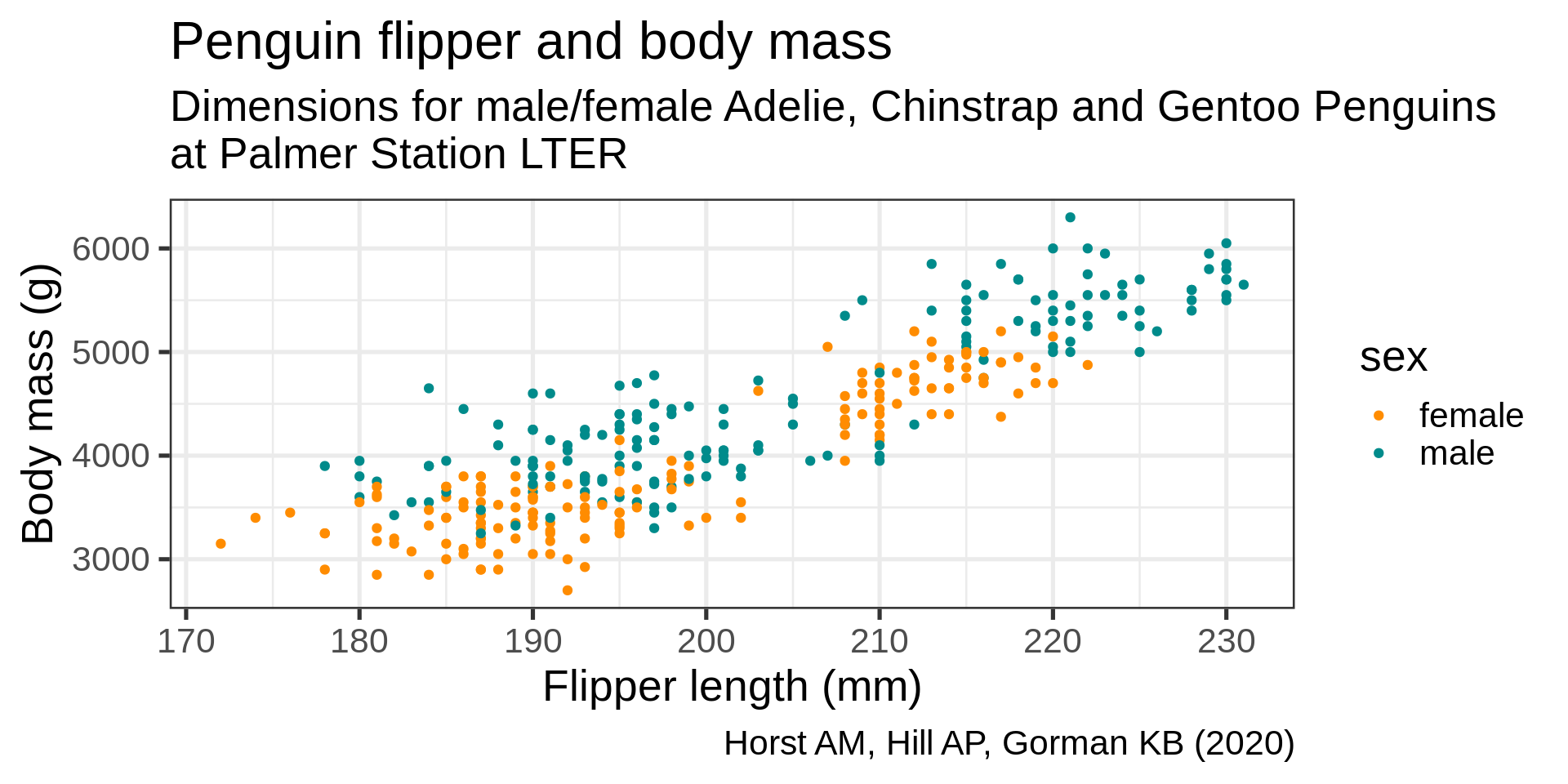
Capabilities in ggplot
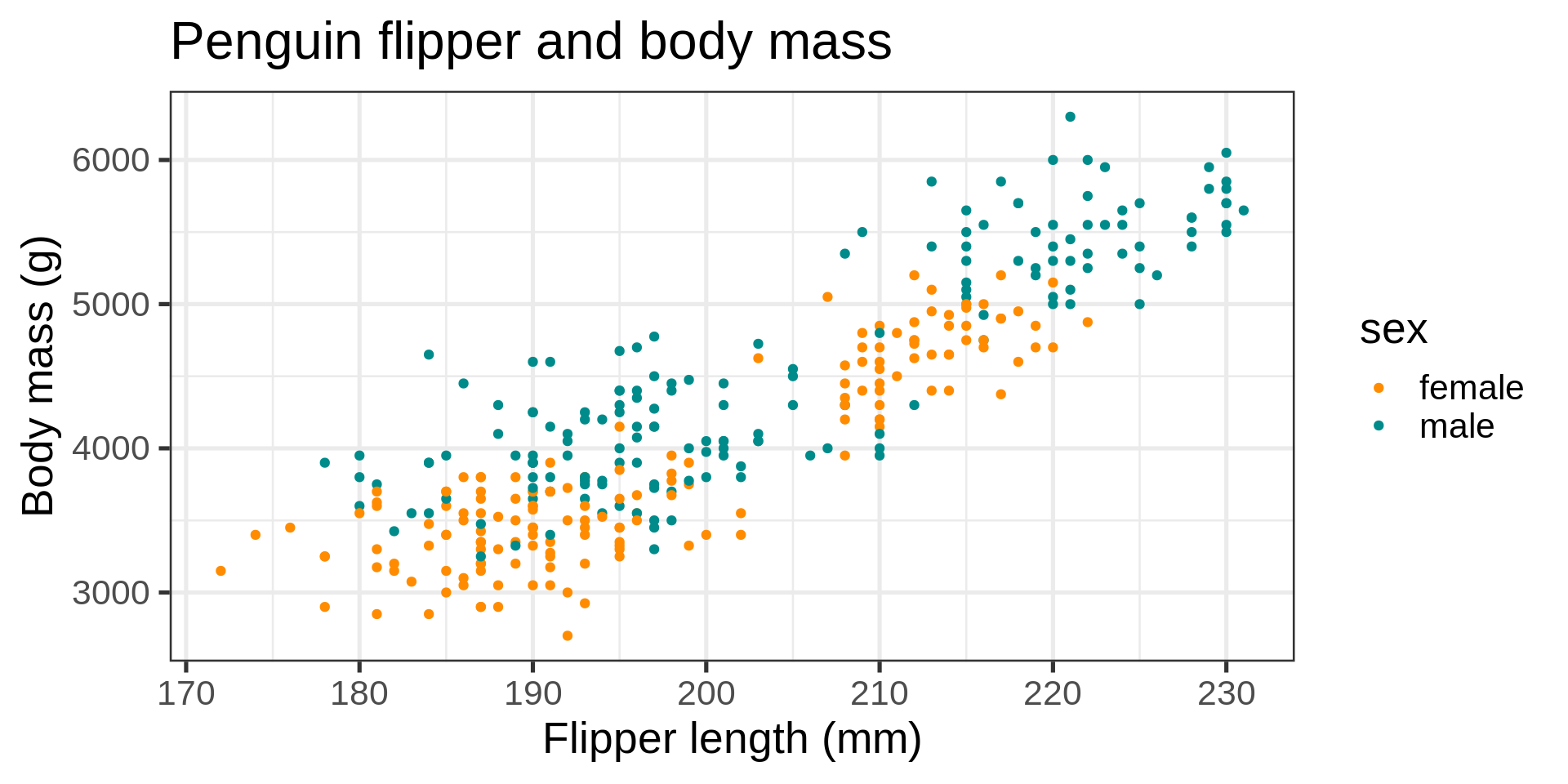
ggplot(data = penguins) +
aes(x = flipper_length_mm,
y = body_mass_g,
color = sex) +
geom_point() +
scale_color_manual(values = c("darkorange", "cyan4"), na.translate = FALSE) +
labs(title = "Penguin flipper and body mass",
caption = "Horst AM, Hill AP, Gorman KB (2020)",
subtitle = description,
x = "Flipper length (mm)",
y = "Body mass (g)",
color = "Penguin sex") +
theme(plot.subtitle = element_text(size = 13),
axis.title = element_text(size = 11)) 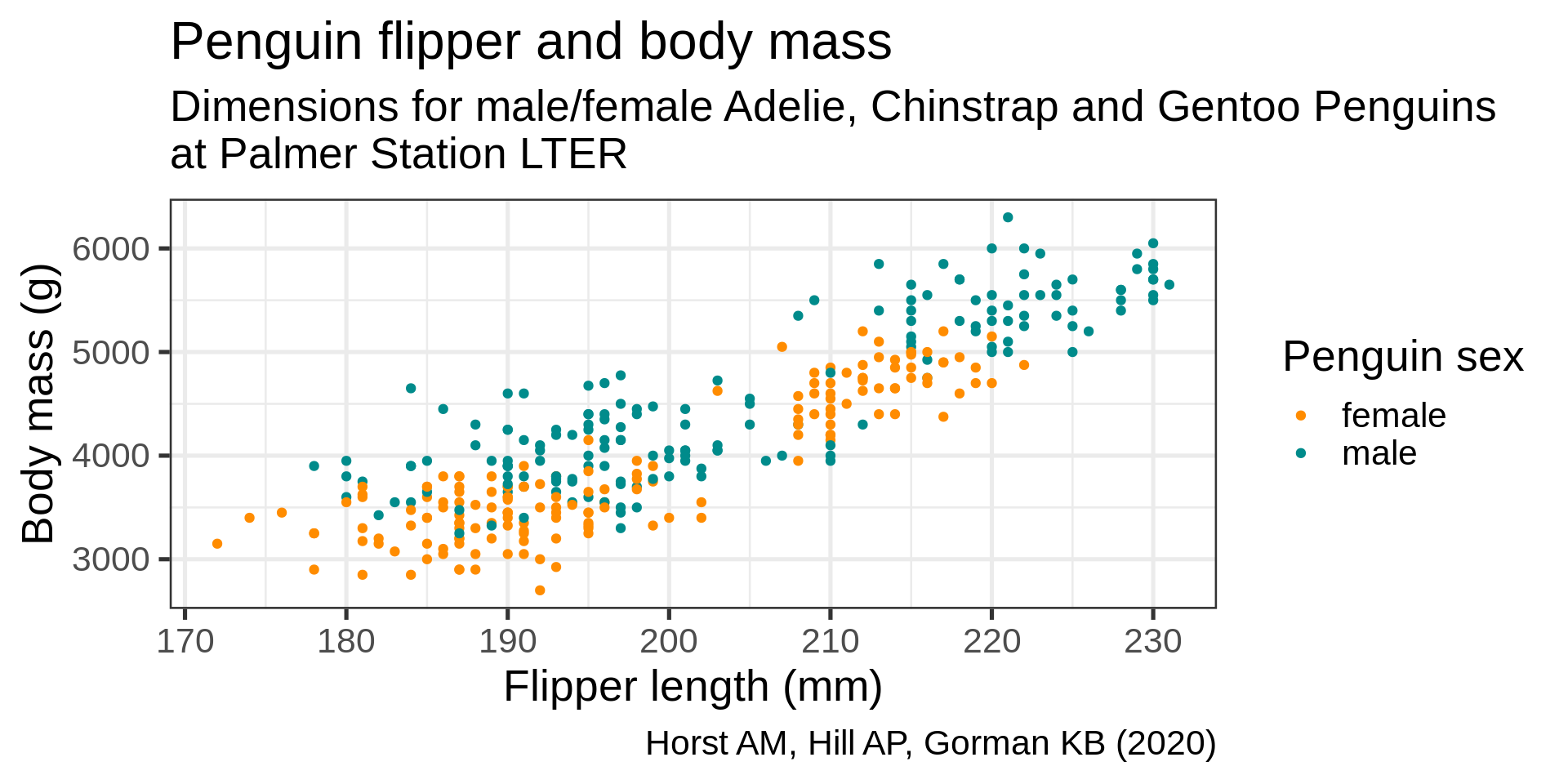
Capabilities in ggplot
ggplot(data = penguins) +
aes(x = flipper_length_mm,
y = body_mass_g,
color = sex) +
geom_point() +
scale_color_manual(values = c("darkorange", "cyan4"), na.translate = FALSE) +
labs(title = "Penguin flipper and body mass",
caption = "Horst AM, Hill AP, Gorman KB (2020)",
subtitle = description,
x = "Flipper length (mm)",
y = "Body mass (g)",
color = "Penguin sex") +
theme(plot.subtitle = element_text(size = 13),
axis.title = element_text(size = 11)) +
theme(legend.position = "bottom",
legend.background = element_rect(fill = "white", color = NA)) 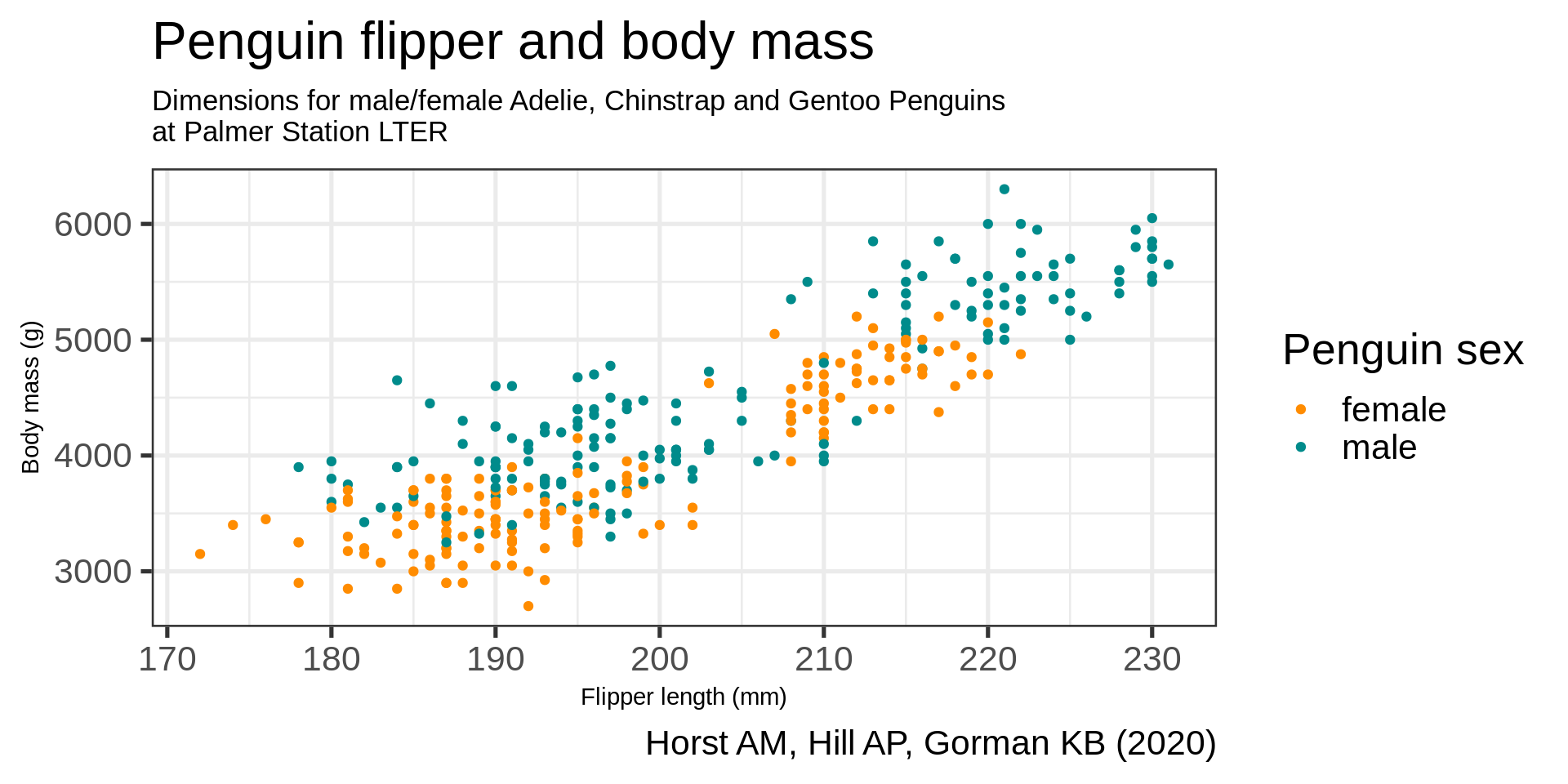
Capabilities in ggplot
ggplot(data = penguins) +
aes(x = flipper_length_mm, y = body_mass_g, color = sex) +
geom_point() +
scale_color_manual(values = c("darkorange", "cyan4"), na.translate = FALSE) +
labs(title = "Penguin flipper and body mass",
caption = "Horst AM, Hill AP, Gorman KB (2020)",
subtitle = description,
x = "Flipper length (mm)",
y = "Body mass (g)",
color = "Penguin sex") +
theme(plot.subtitle = element_text(size = 13),
axis.title = element_text(size = 11)) +
theme(legend.position = "bottom",
legend.background = element_rect(fill = "white", color = NA)) +
theme(plot.caption = element_text(hjust = 0, face = "italic"),
plot.caption.position = "plot") +
facet_wrap(vars(species))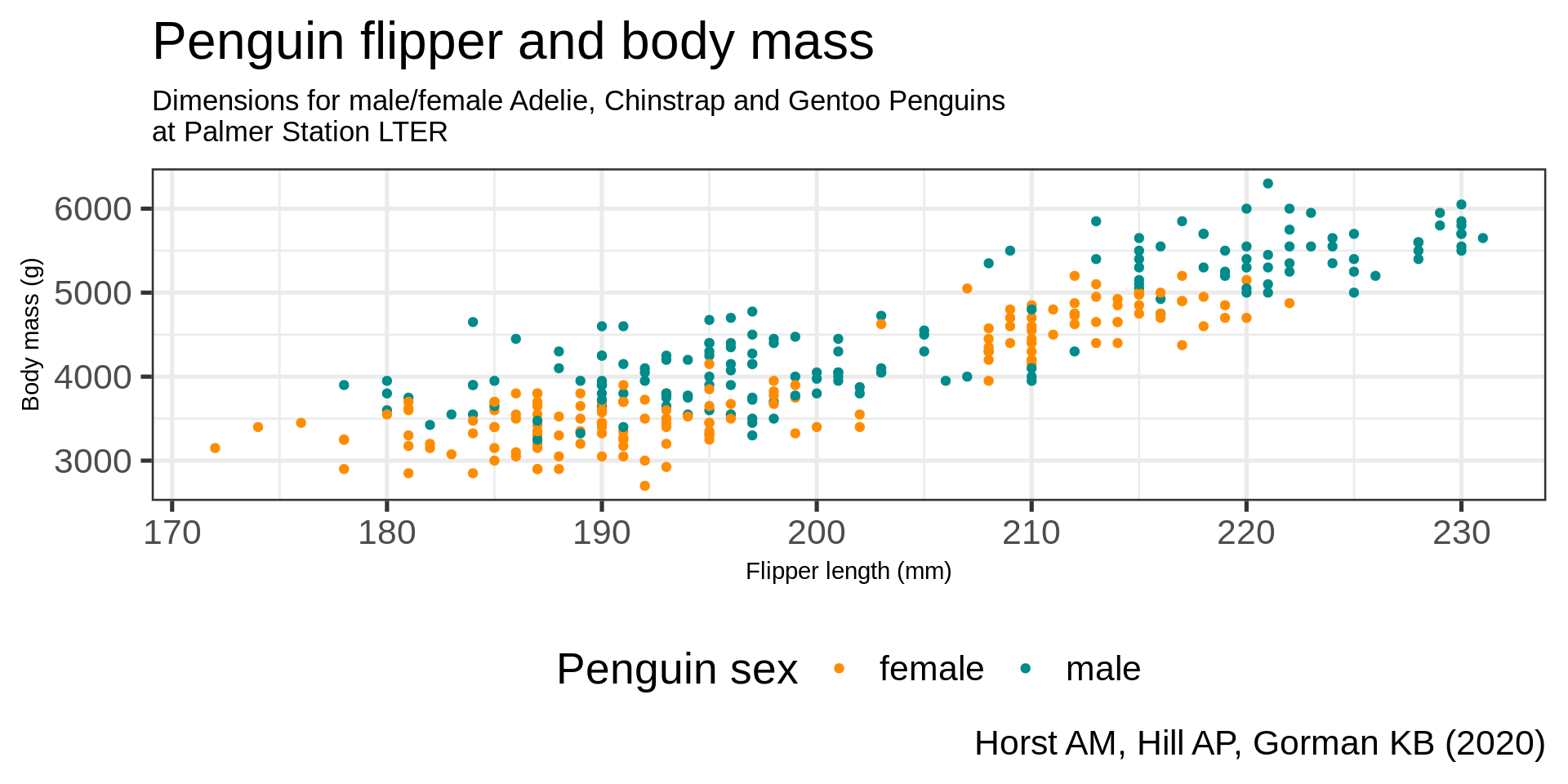
Capabilities in ggplot
ggplot(data = penguins) +
aes(x = flipper_length_mm, y = body_mass_g, color = sex) +
geom_point() +
scale_color_manual(values = c("darkorange", "cyan4"), na.translate = FALSE) +
labs(title = "Penguin flipper and body mass",
caption = "Horst AM, Hill AP, Gorman KB (2020)",
subtitle = description,
x = "Flipper length (mm)",
y = "Body mass (g)",
color = "Penguin sex") +
theme(plot.subtitle = element_text(size = 13),
axis.title = element_text(size = 11)) +
theme(legend.position = "bottom",
legend.background = element_rect(fill = "white", color = NA)) +
theme(plot.caption = element_text(hjust = 0, face = "italic"),
plot.caption.position = "plot") +
facet_wrap(vars(species)) +
scale_x_continuous(guide = guide_axis(n.dodge = 2)) +
scale_y_continuous(labels = scales::label_comma())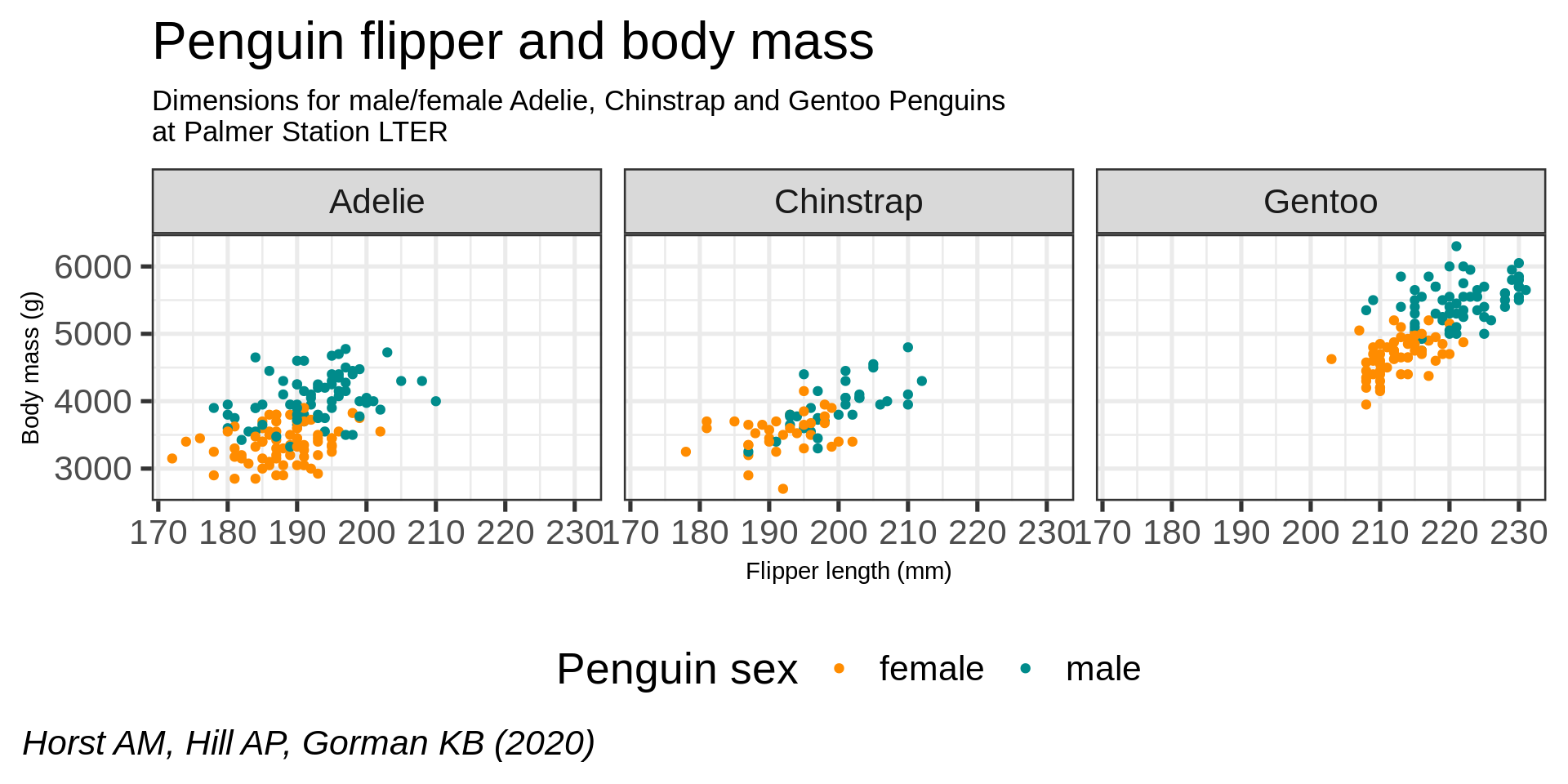
Geometric objects define the plot type to be drawn
geom_point()
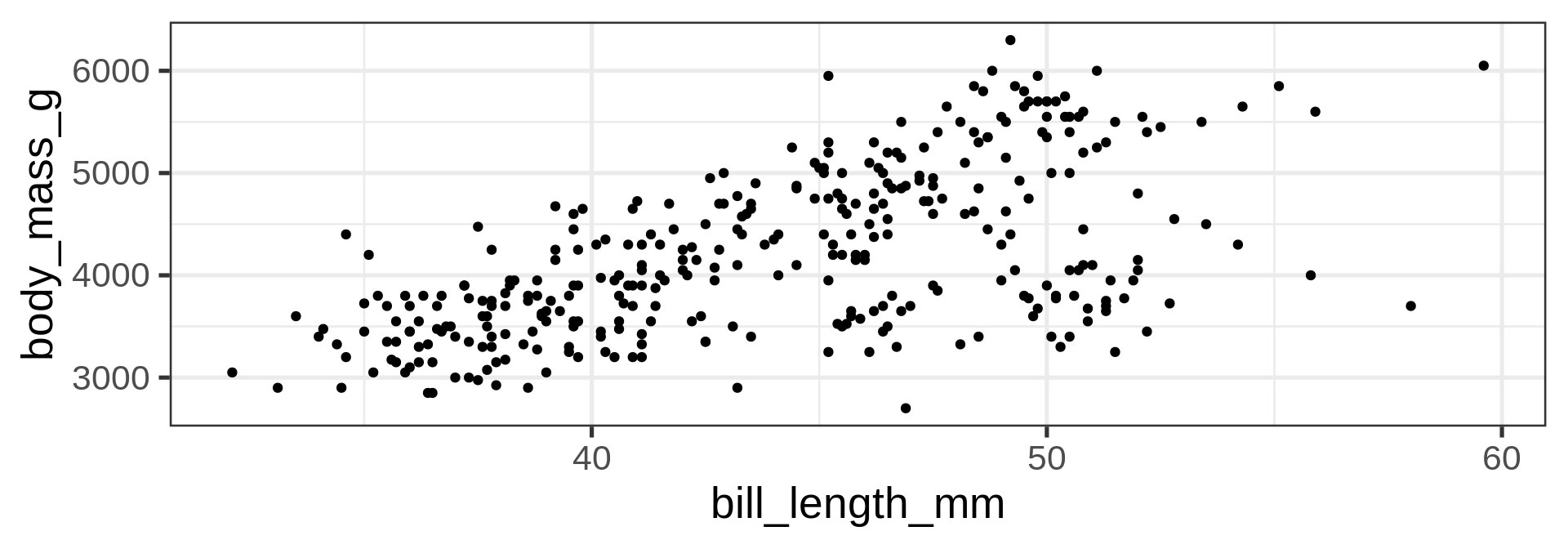
geom_line()
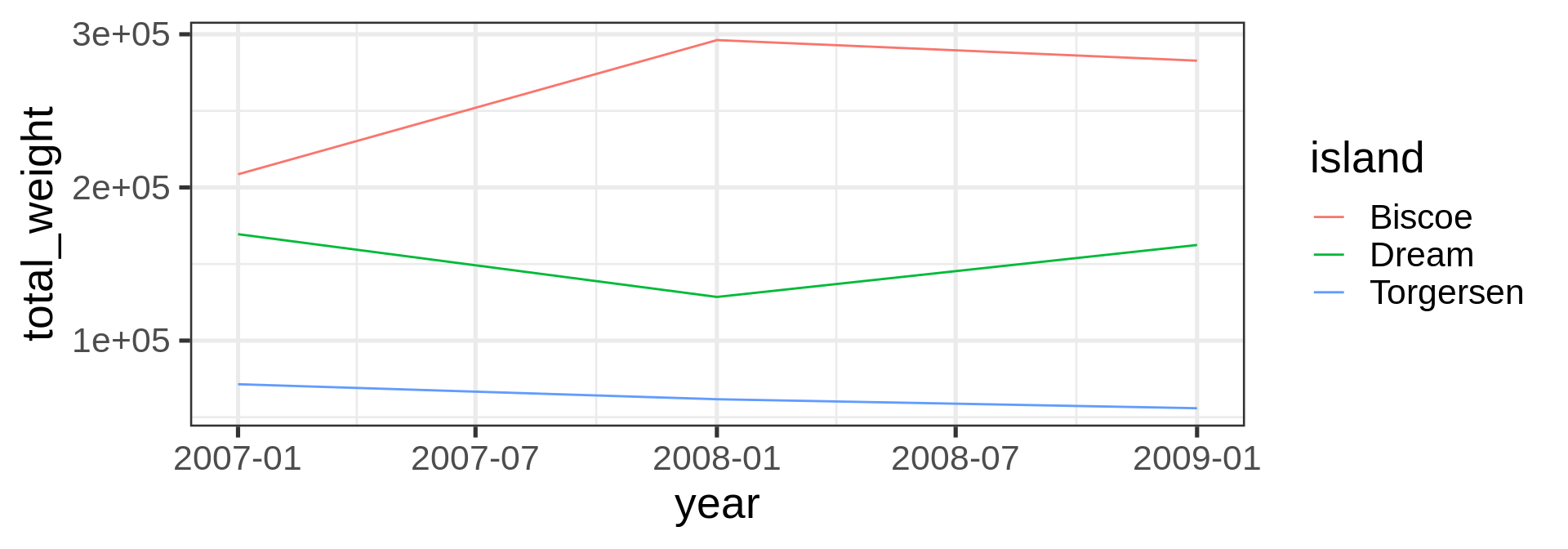
geom_bar()
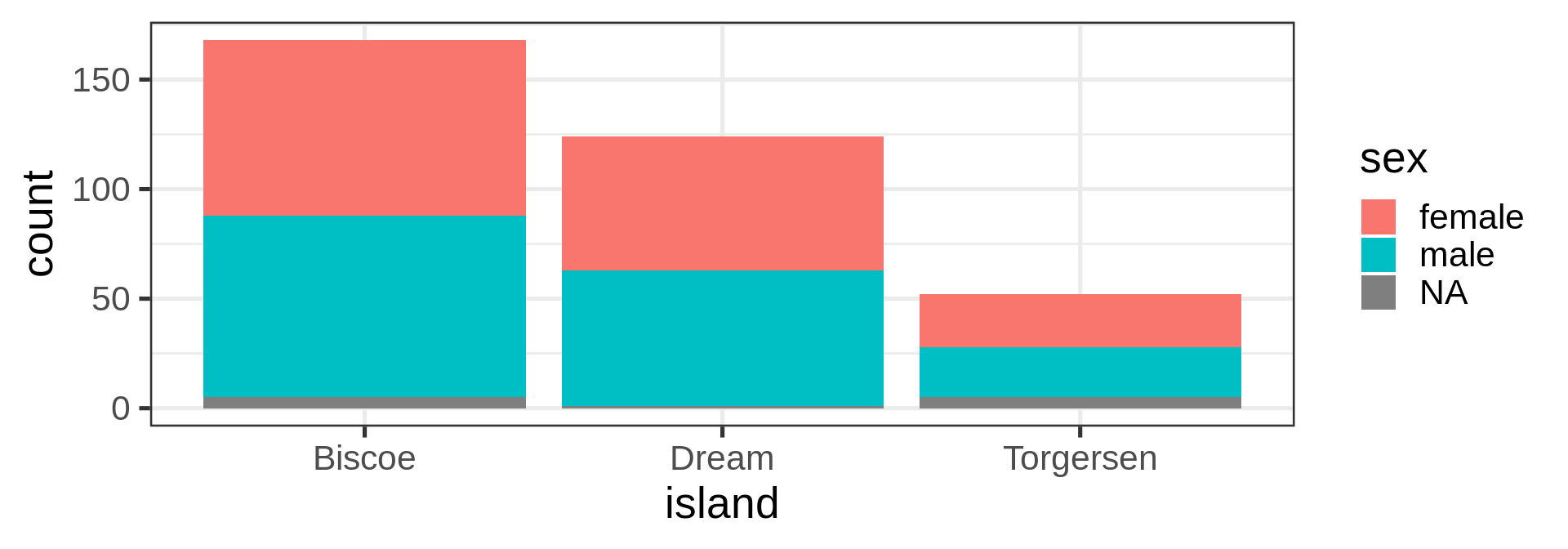
geom_violin()
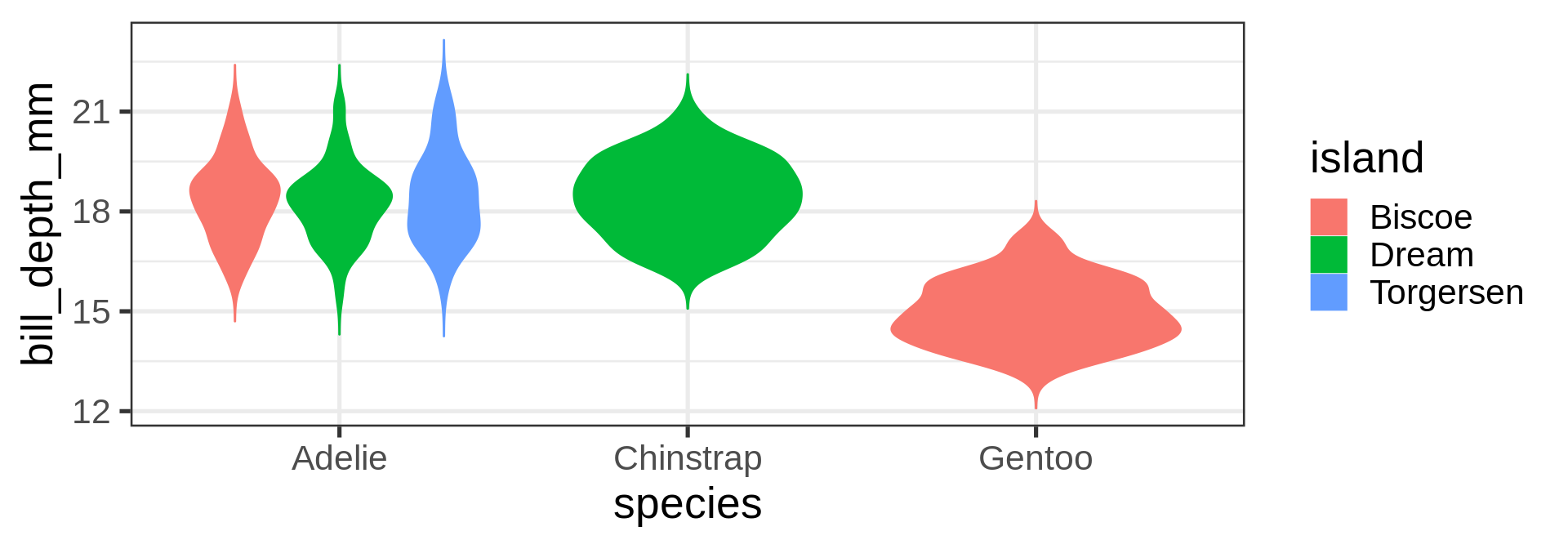
geom_histogram()
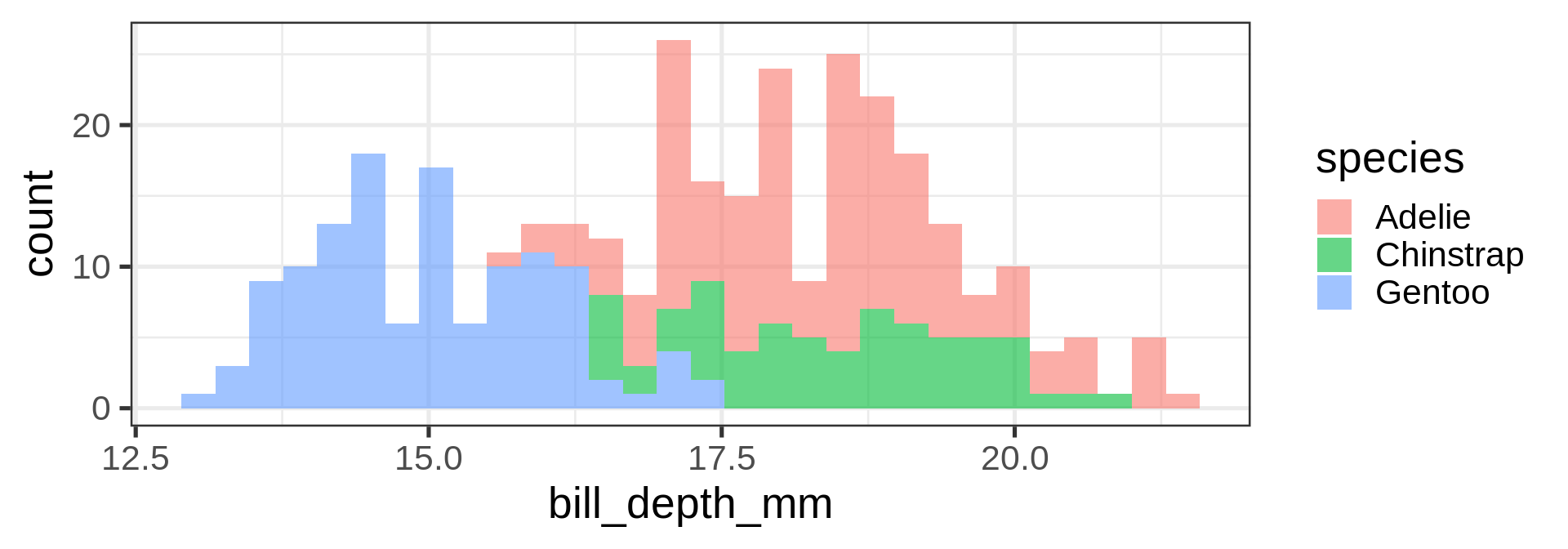
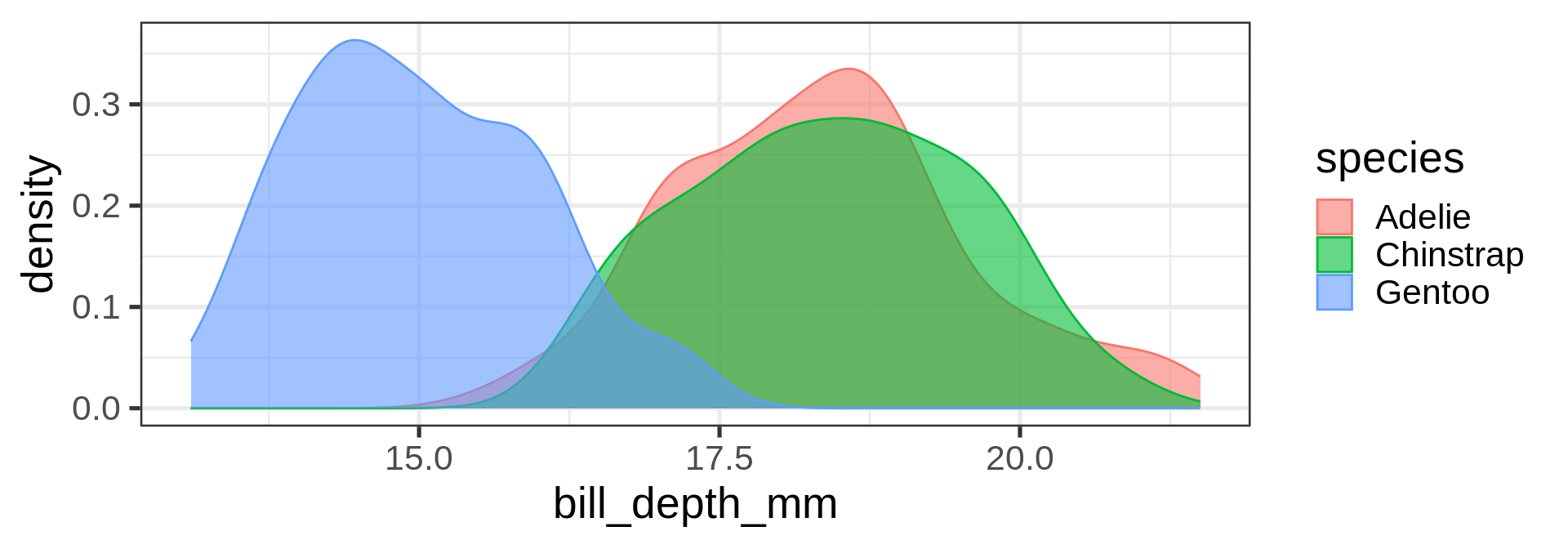
geom_density()
Have a look at the cheatsheet or the documentation for more possibilities.
Layers
Core layers
Other layers
They are present, it works because they have sensible default:
- Theme is
theme_grey - Coordinate is
cartesian - Statistic is
identity - Facets are
disabled
Three layers are sufficient
- Data
- Aesthetics mapping data to plot component
- Geometry at least one
Your first plot
My first plot
Mapping aesthetics
Requirements
aes()map columns/variables data to aesthetics- Specific geometries (
geom) have different expectations:- univariate, one x or y for flipped axes
- bivariate, x and y like scatterplot
- Continuous or Discrete variables
- Continuous for color ➡️ gradient
- Discrete for color ➡️ qualitative
NB: mapping = and data = are often skipped.
- Same as previous slide
- Without
colourmapping
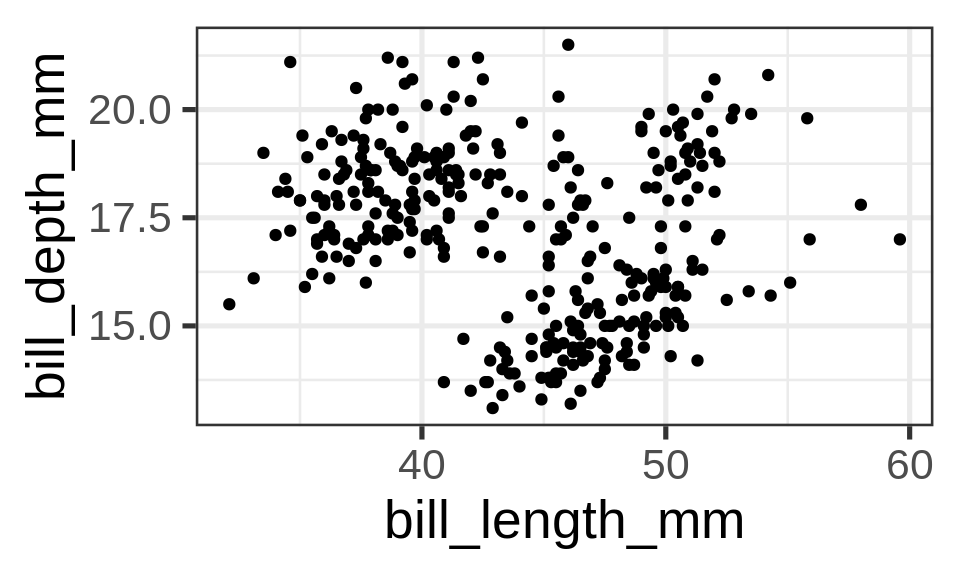
Unmapped parameters
Mapped parameters
Mapped parameters require two conditions:
- Being defined inside the aesthetics
aes() - Refer to one of the column data, here: mistake
Error in FUN(X[[i]], ...): object 'country' not found- Passing the unknown column as
stringas a different effect:
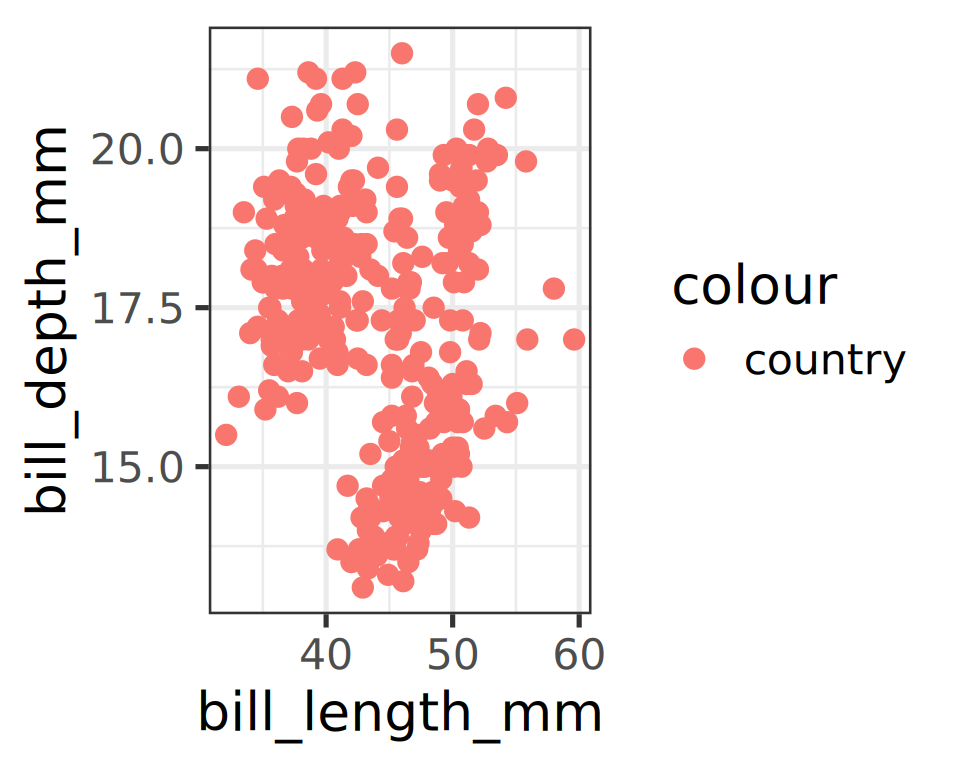
Mapping rules
In aes()
refer to a valid column.
Mapping aesthetics correctly
In aes() and refer to a data column
Why no string for mapping?
Inheritance of arguments across layers
Compare the code and the results
Note
aestheticsinggplot()are passed on to allgeometries.
Note
aestheticsingeom_*()are specific (and can overwrite inherited)
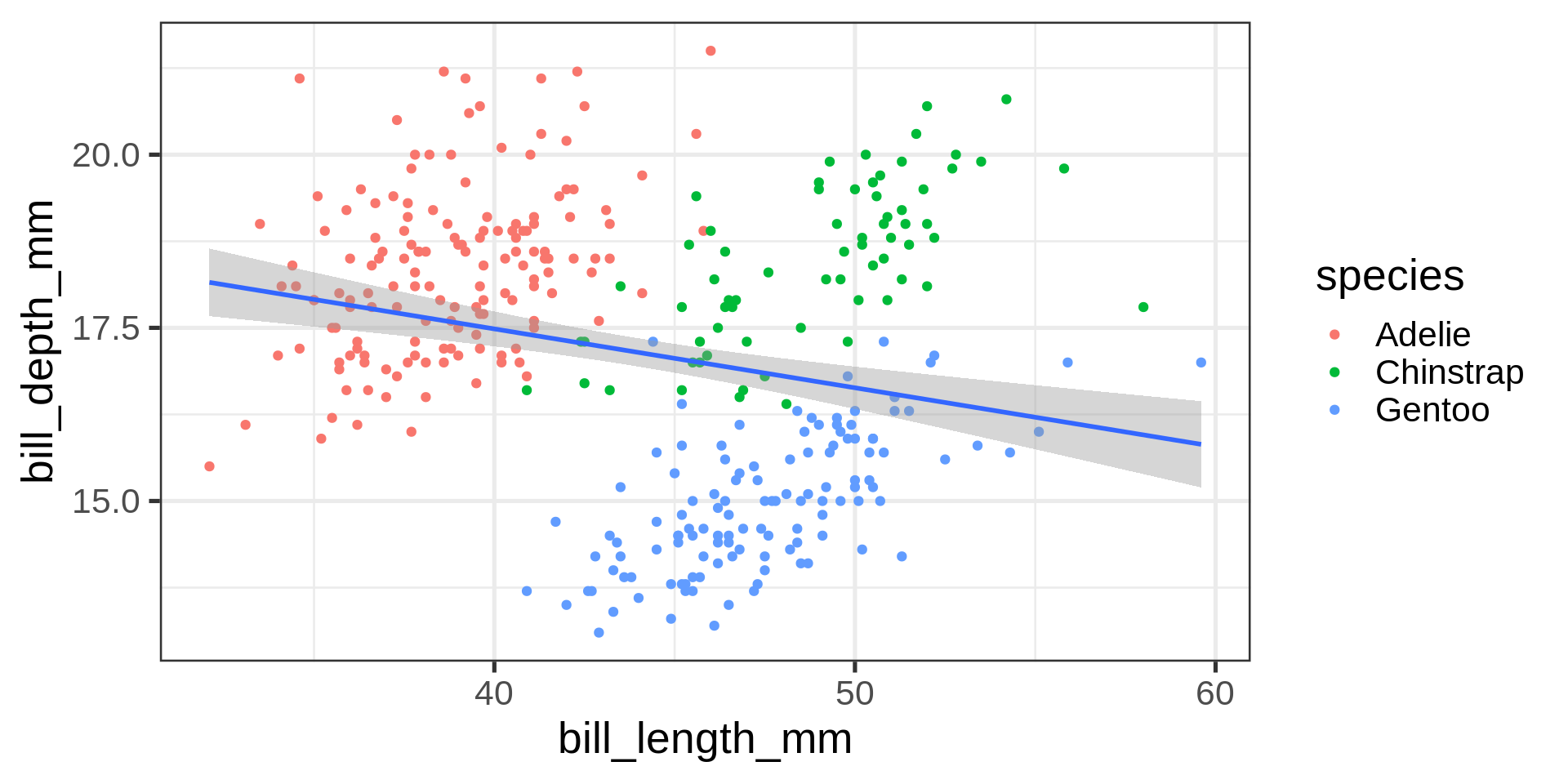
Simpson’s paradox
Statistical correlation depending on stratification.
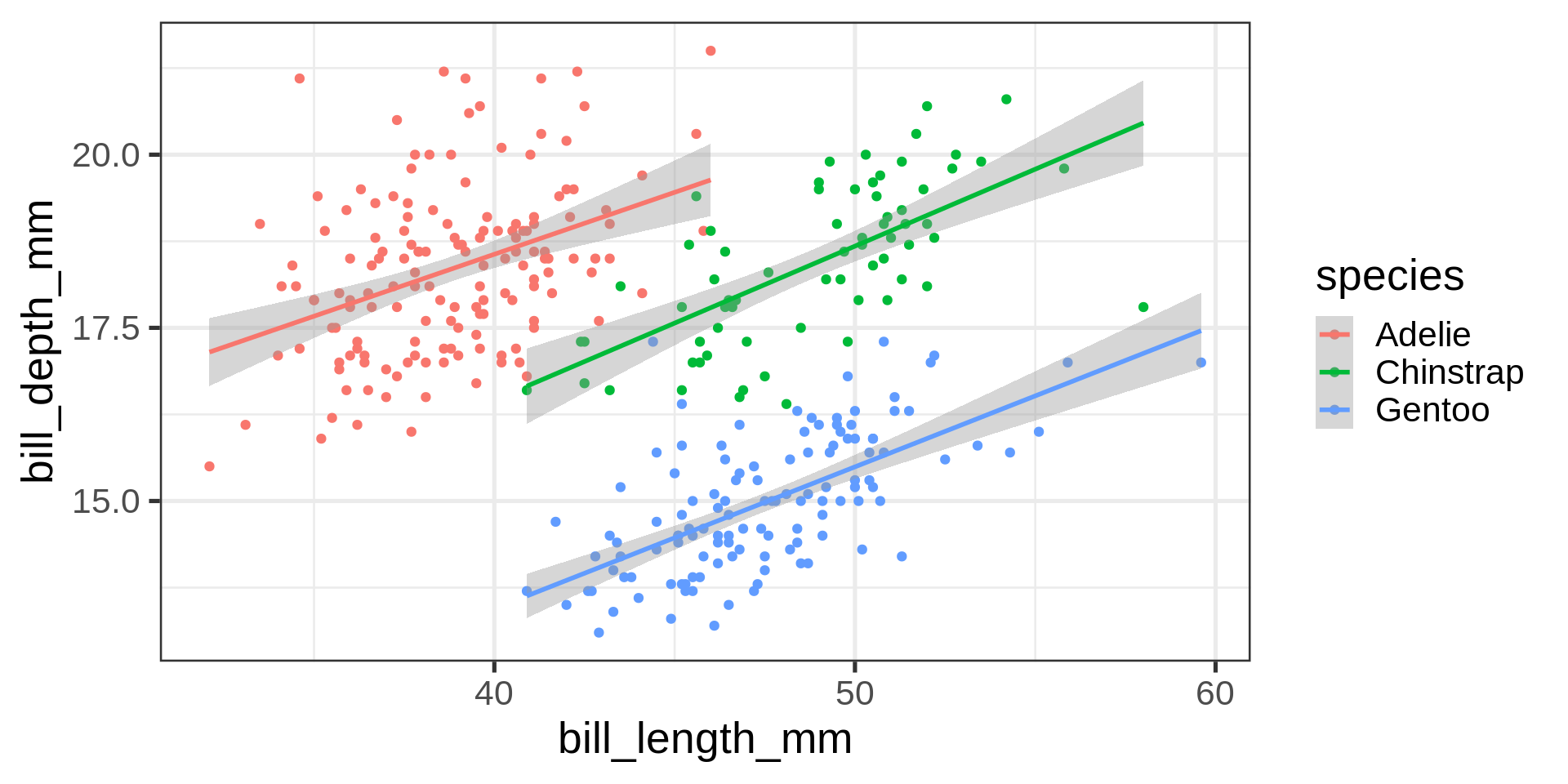
Complete data set $R^2 = $ 0.055
Your turn!
- Use the classroom practical.
- Install
palmerpenguinspackage if you haven’t yet - Use the
penguinsdata set and plotbill_length_mm,bill_depth_mmandspecies. - Map the variable
islandto the aestheticsshape. - Add a regression line using a linear model.
- All dots (circles / triangles / squares) with:
- A size of
5 - A transparency of 30% (
alpha = 0.7)
- A size of
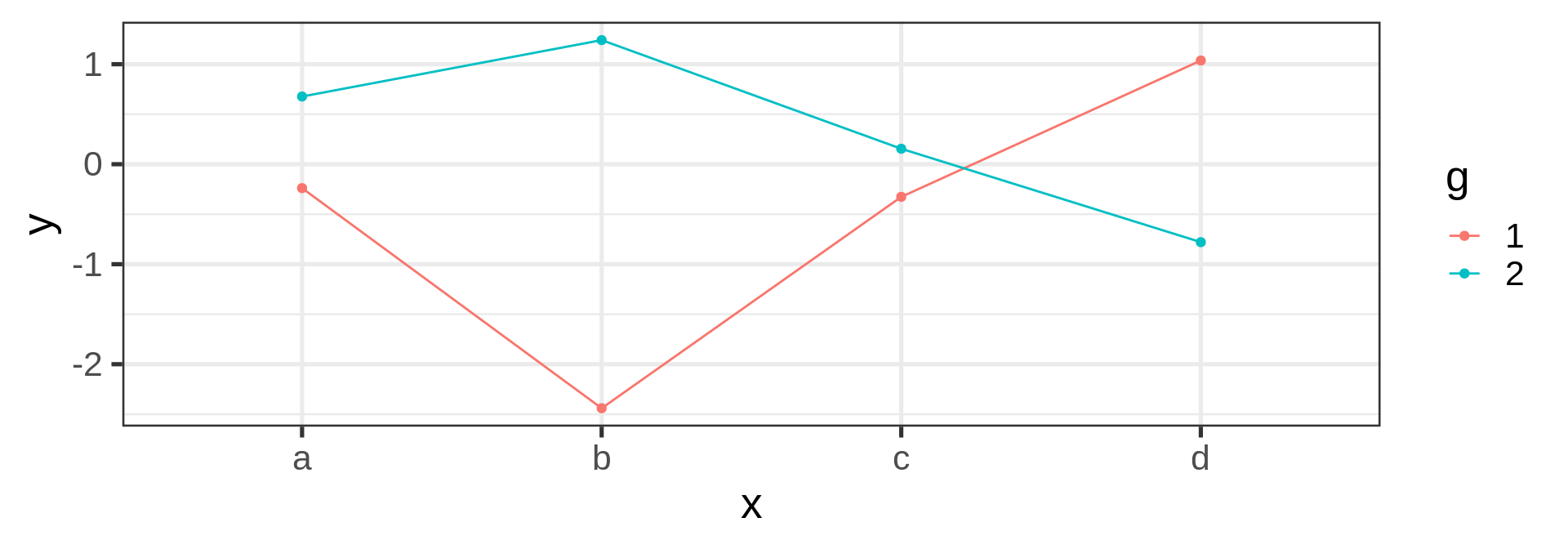
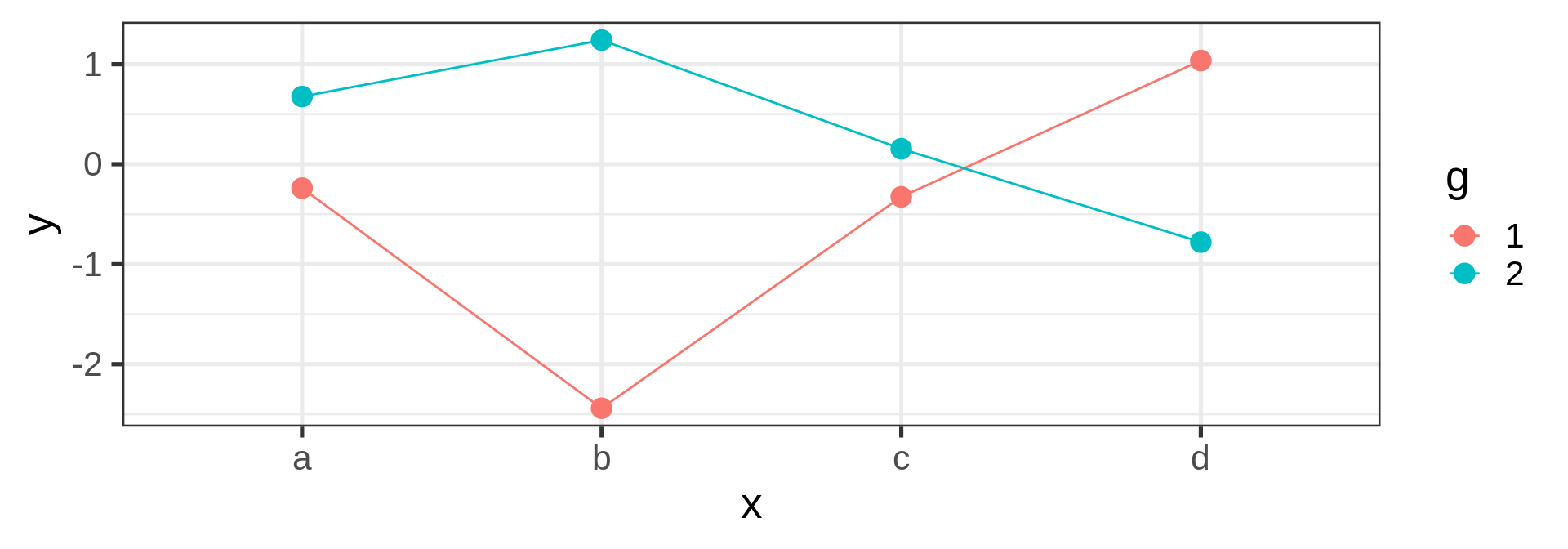
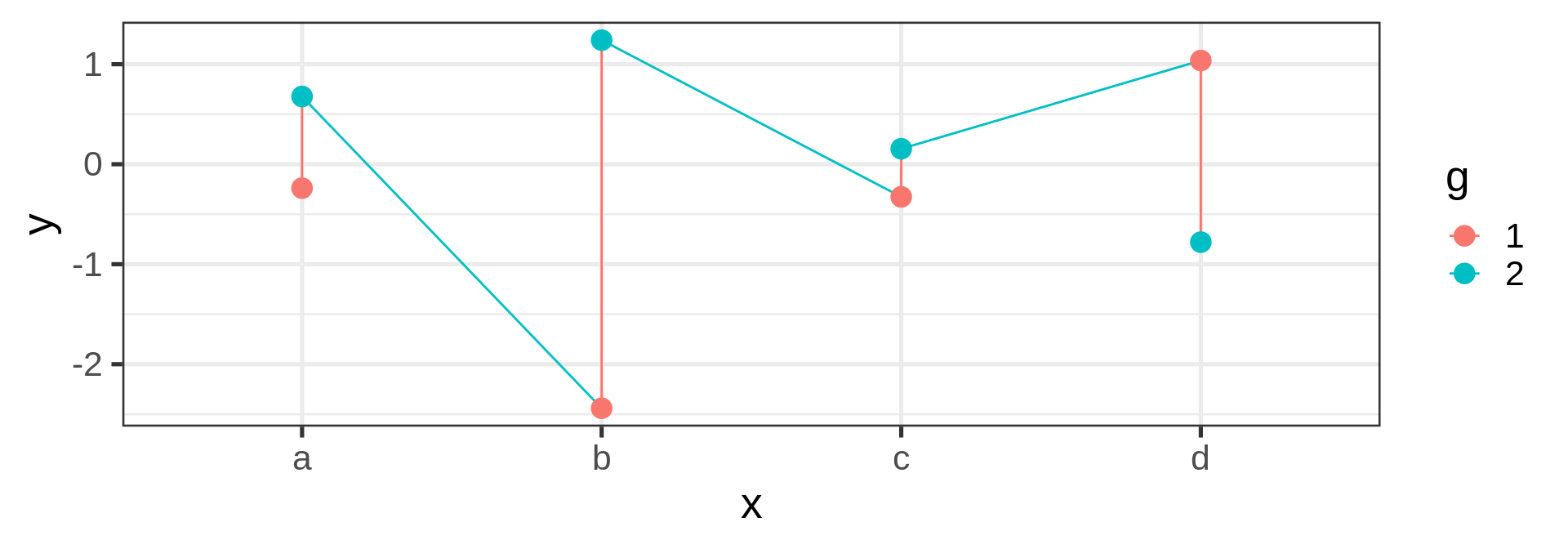
Joining observations
Suppose we want to connect dots by colors
Warning
Should be the job of geom_line()
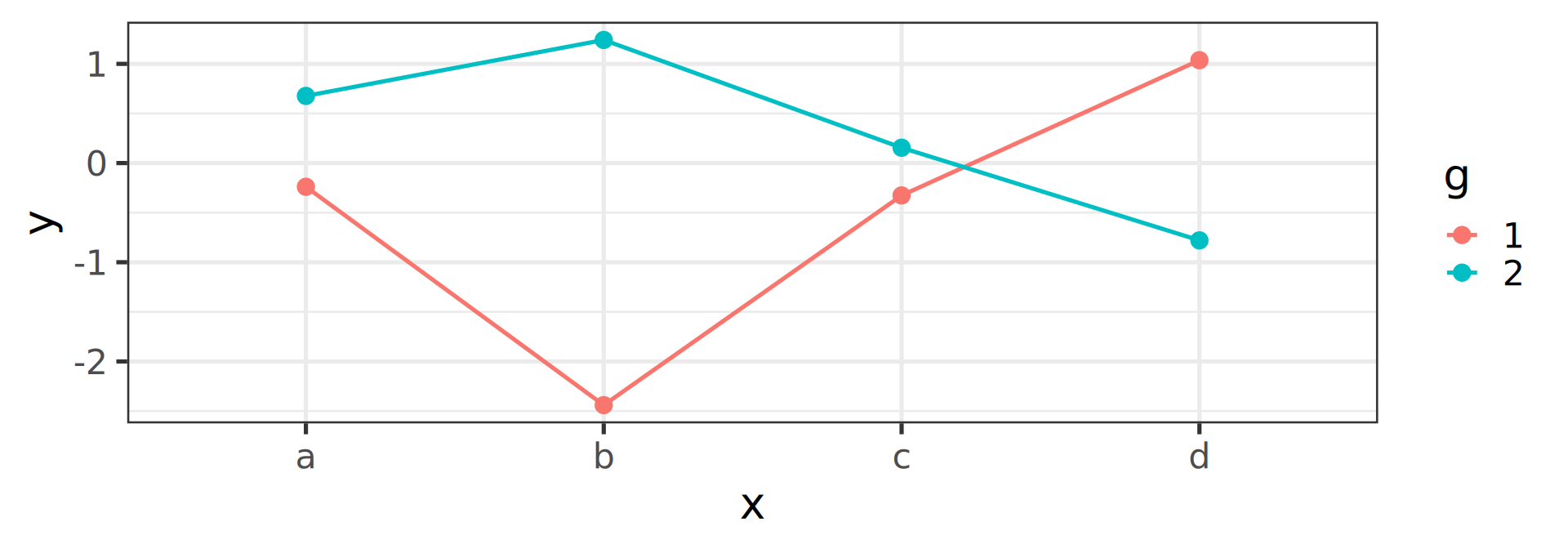
Invisible aesthetic: grouping
Source: koshske blog: geom_line() doesn’t draw lines
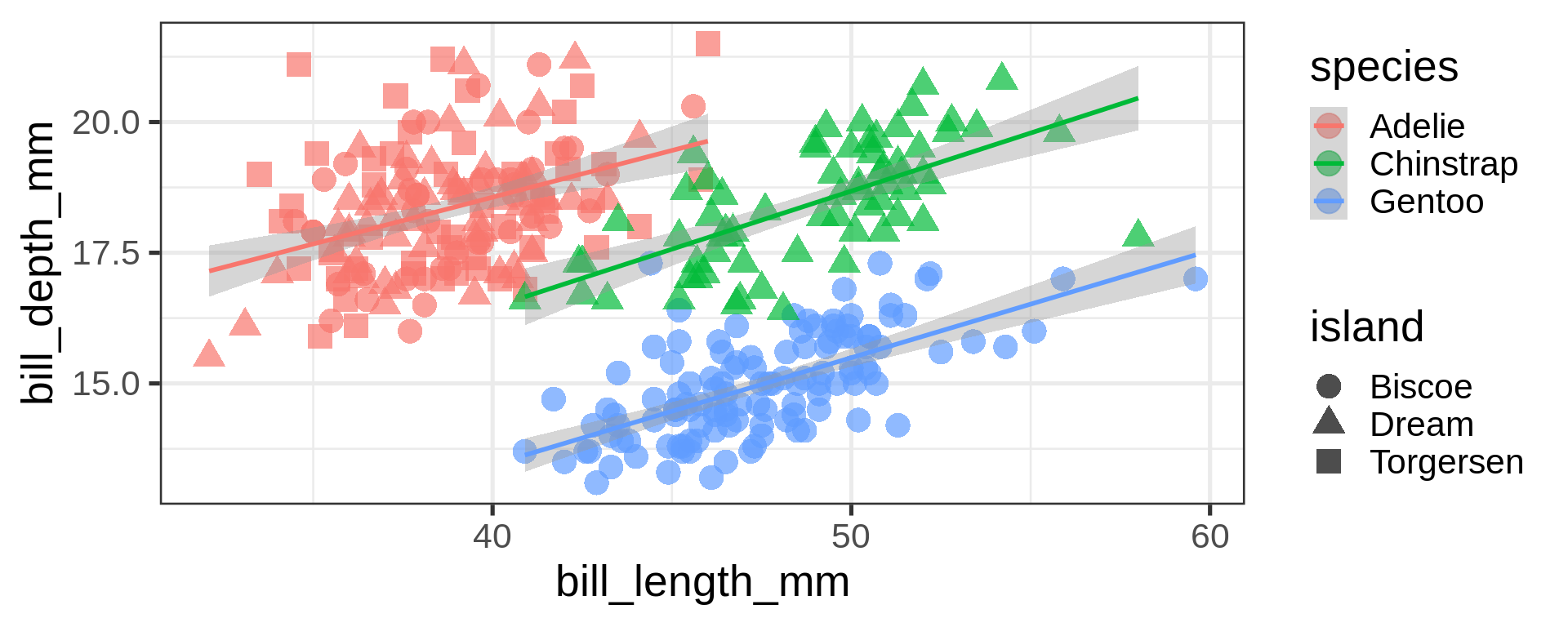
Labels
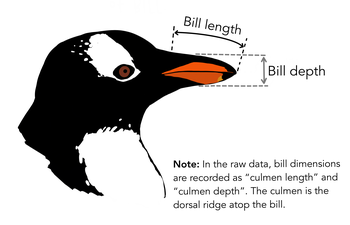
ggplot(penguins,
aes(x = bill_length_mm,
y = bill_depth_mm,
shape = island,
colour = species)) +
geom_point() +
geom_smooth(method = "lm",
formula = 'y ~ x') +
labs(title = "Bill ratios of Palmer penguins",
caption = "Horst AM, Hill AP, Gorman KB (2020)",
subtitle = "Split per species / island",
shape = "Islands",
x = "cumen length (mm)",
y = "cumen depth (mm)")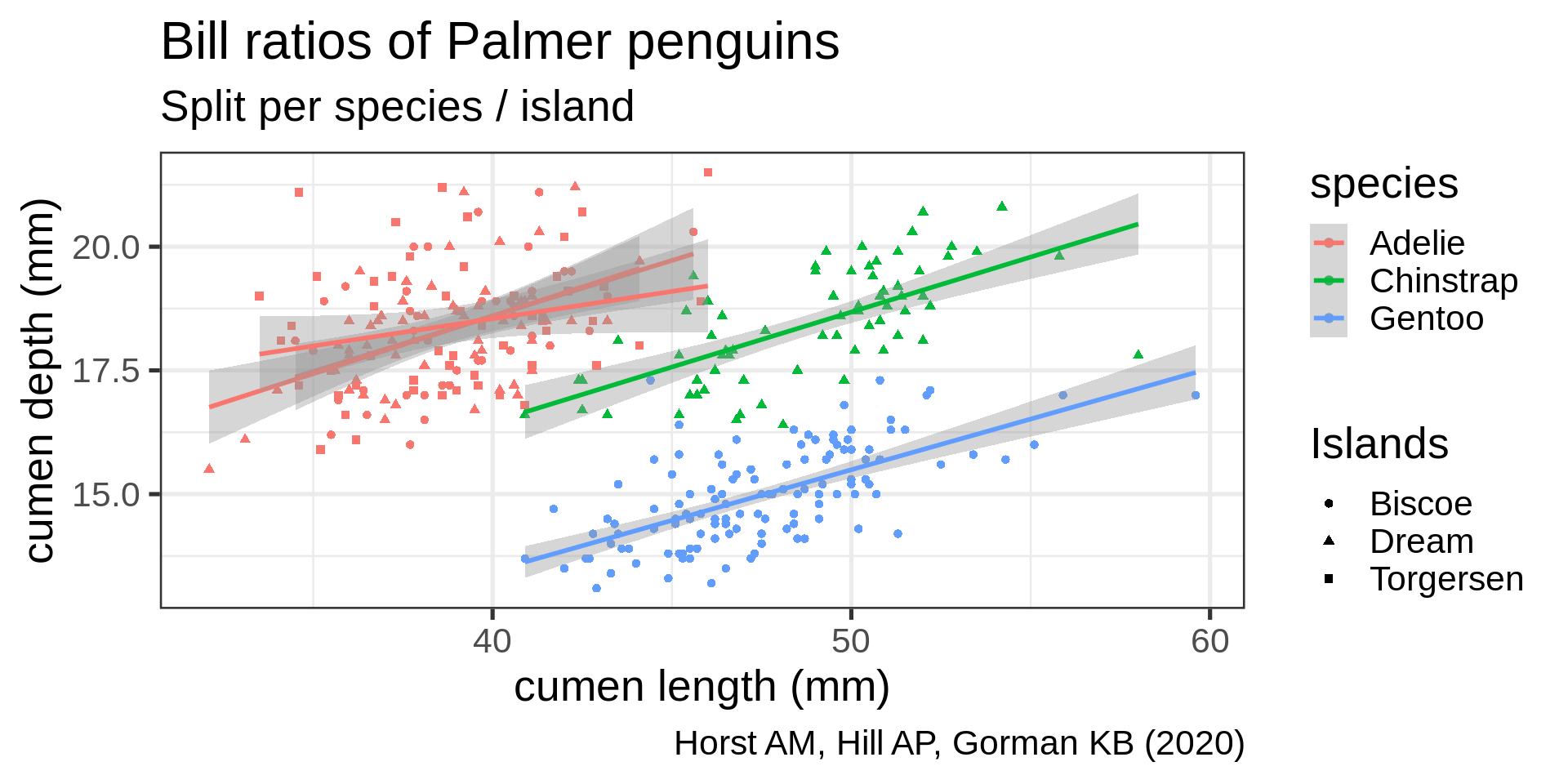
Statistics / geometries are interchangeable
Let ggplot2 doing the stat for you
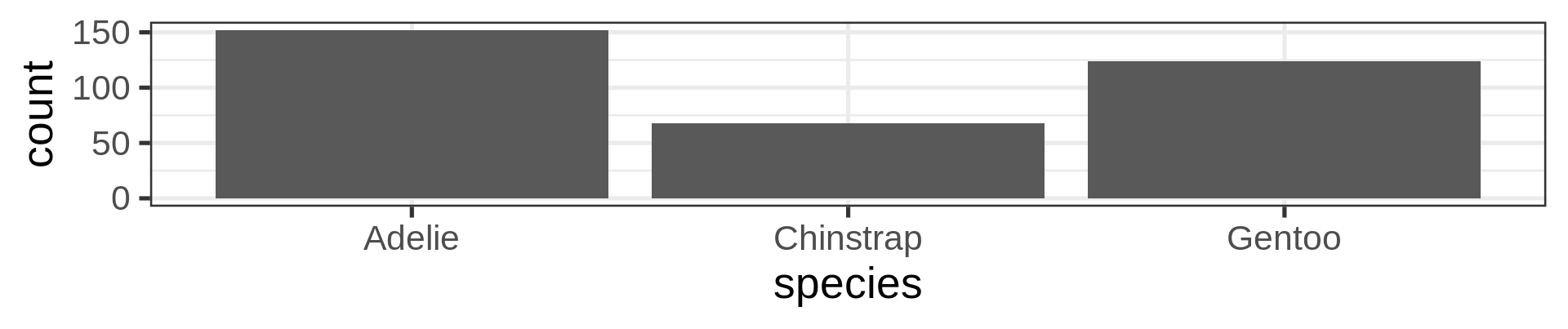
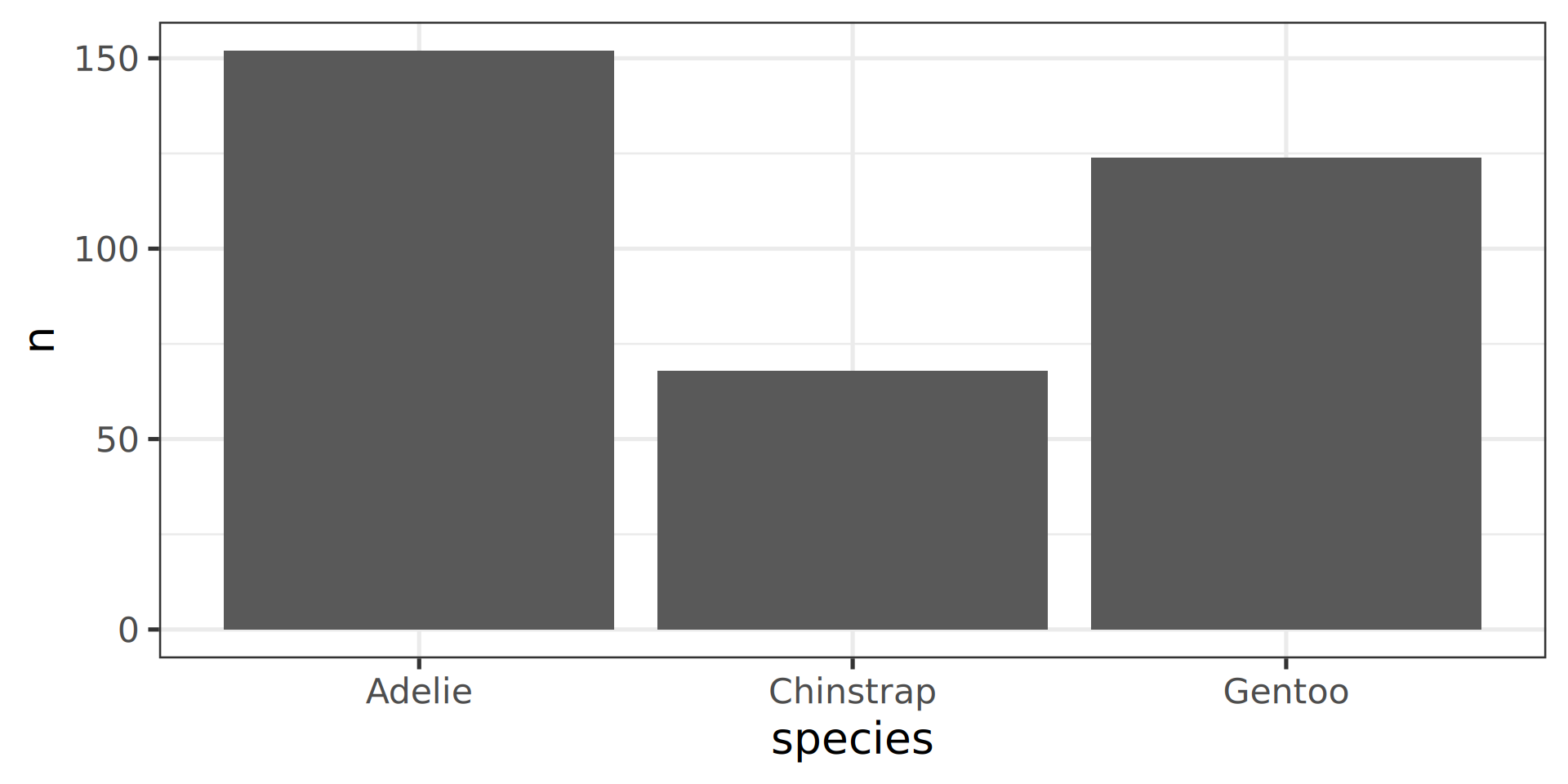
Count yourself geom_col()
The stat() function allows computation, like proportions
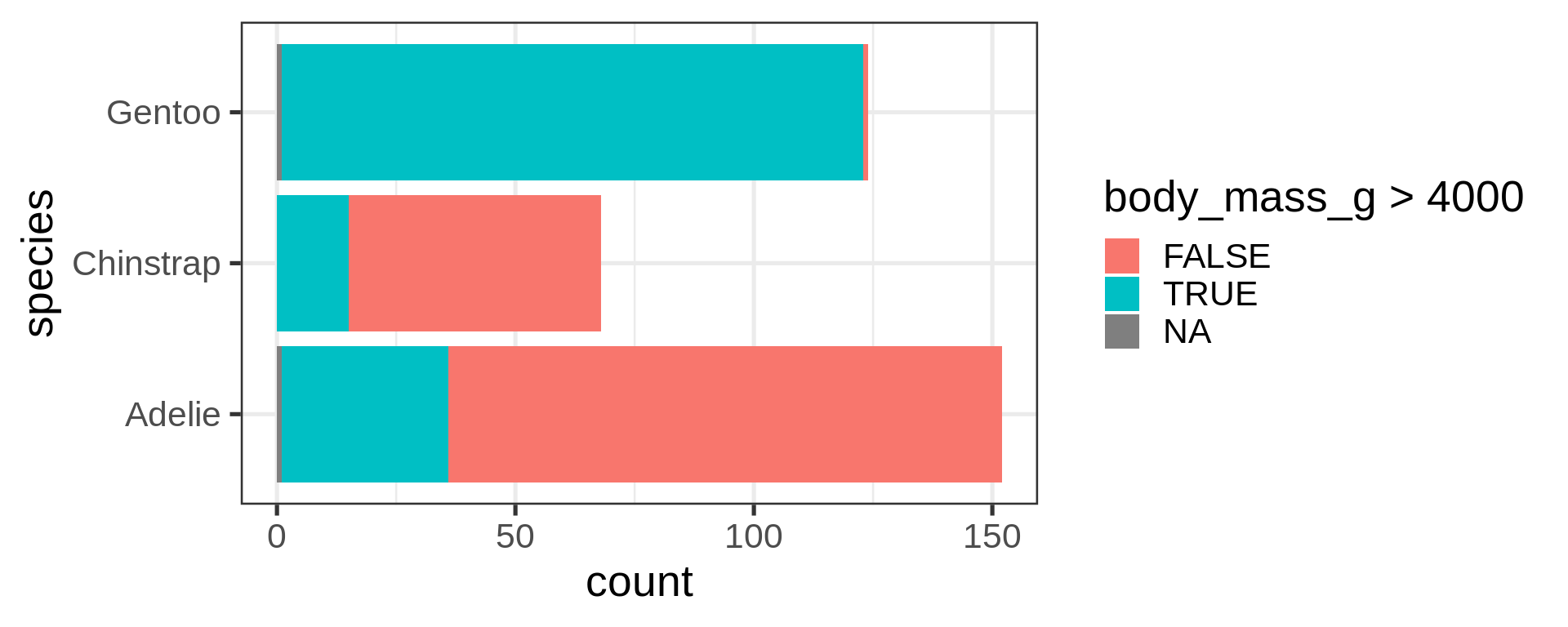
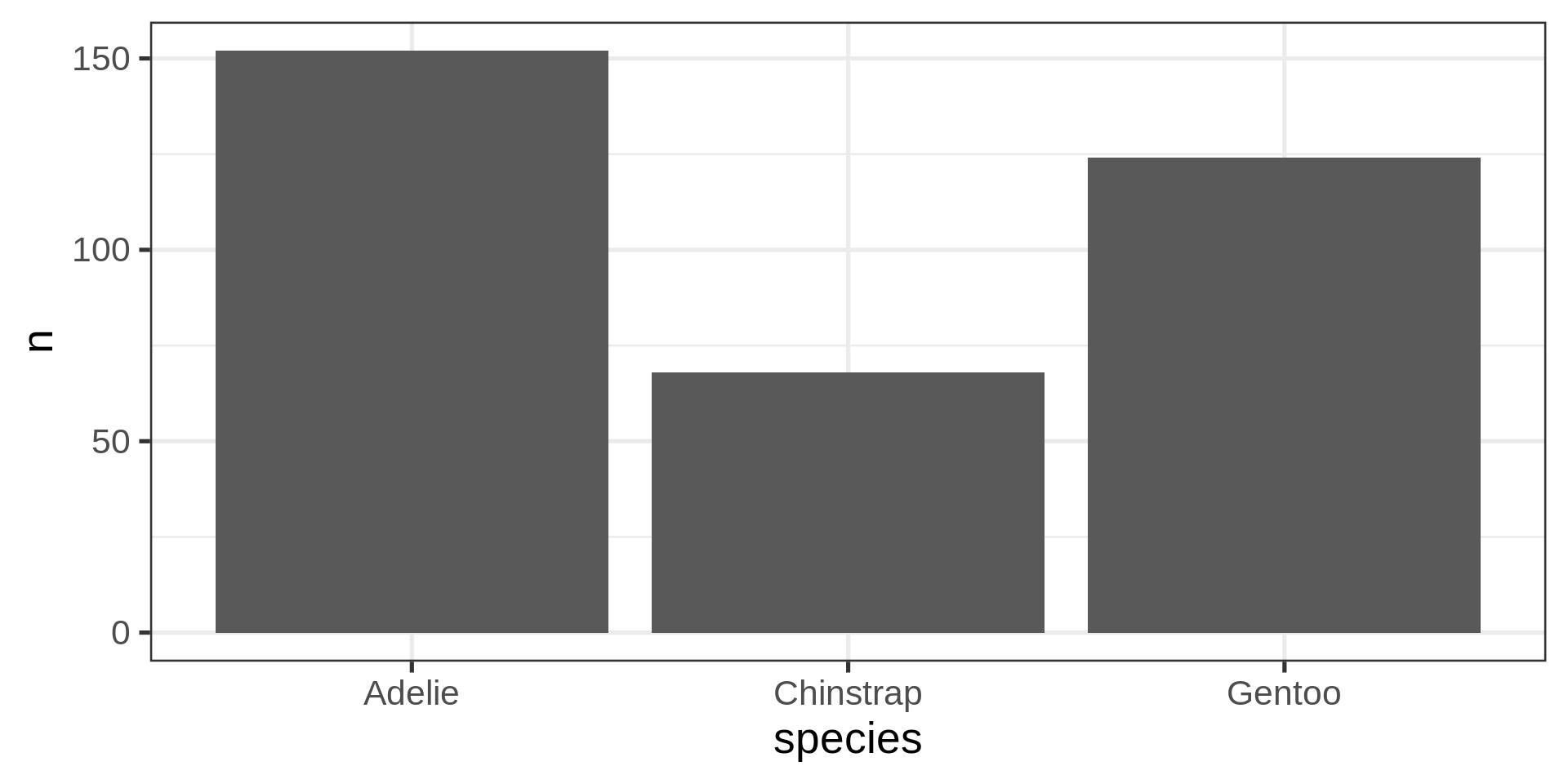
Classic counting
See list in help pages

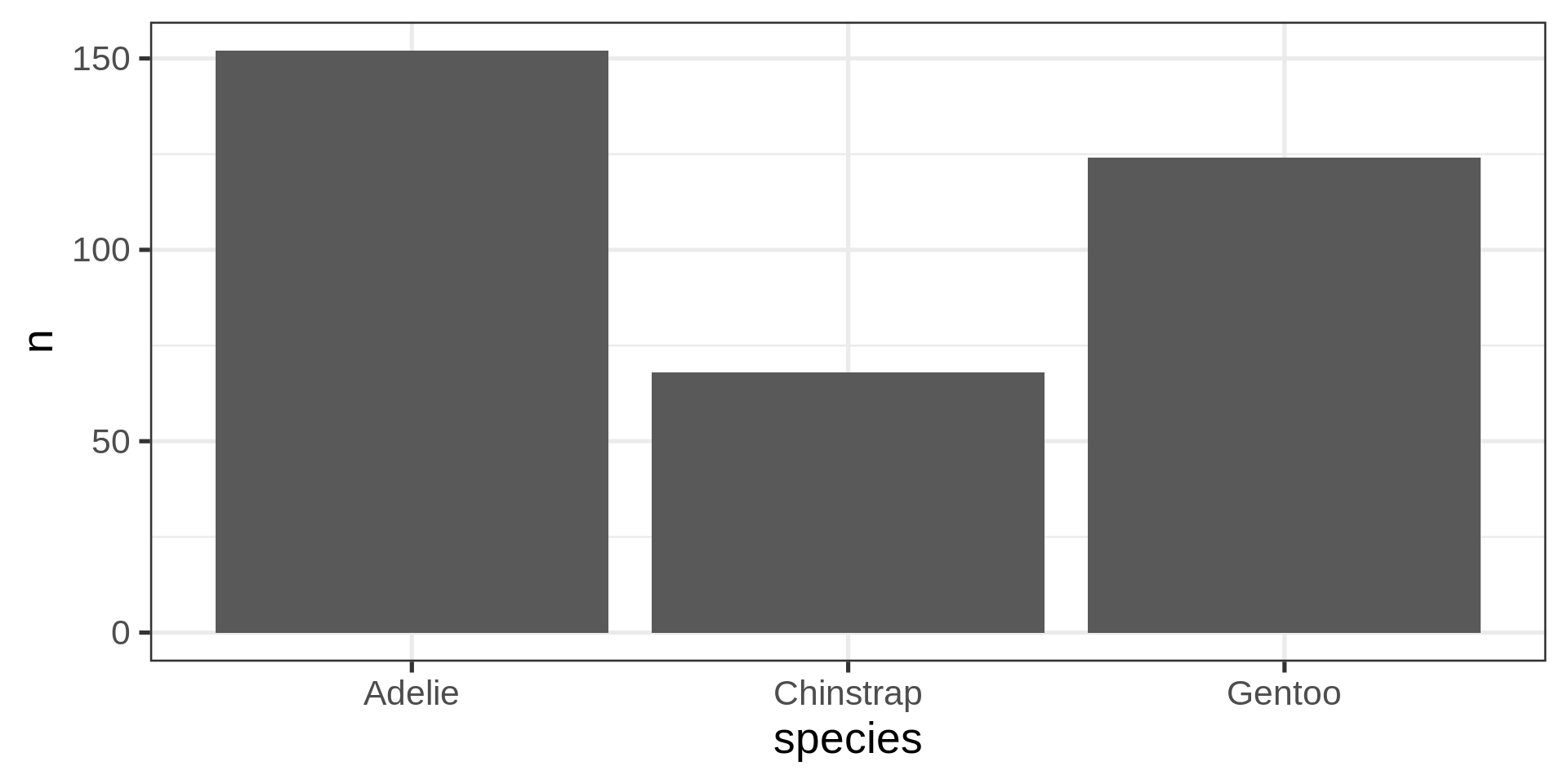
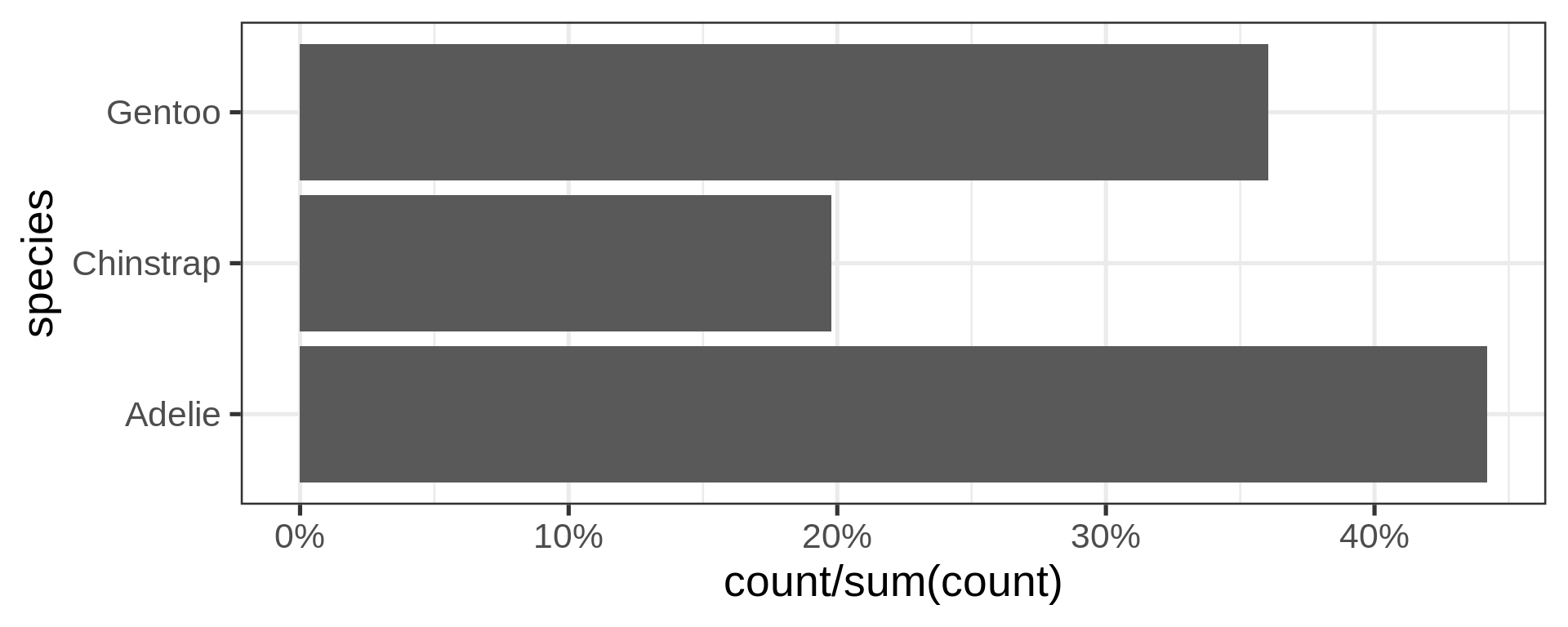
- Now compute proportions
- Bonus: get
xscale in%usingscales
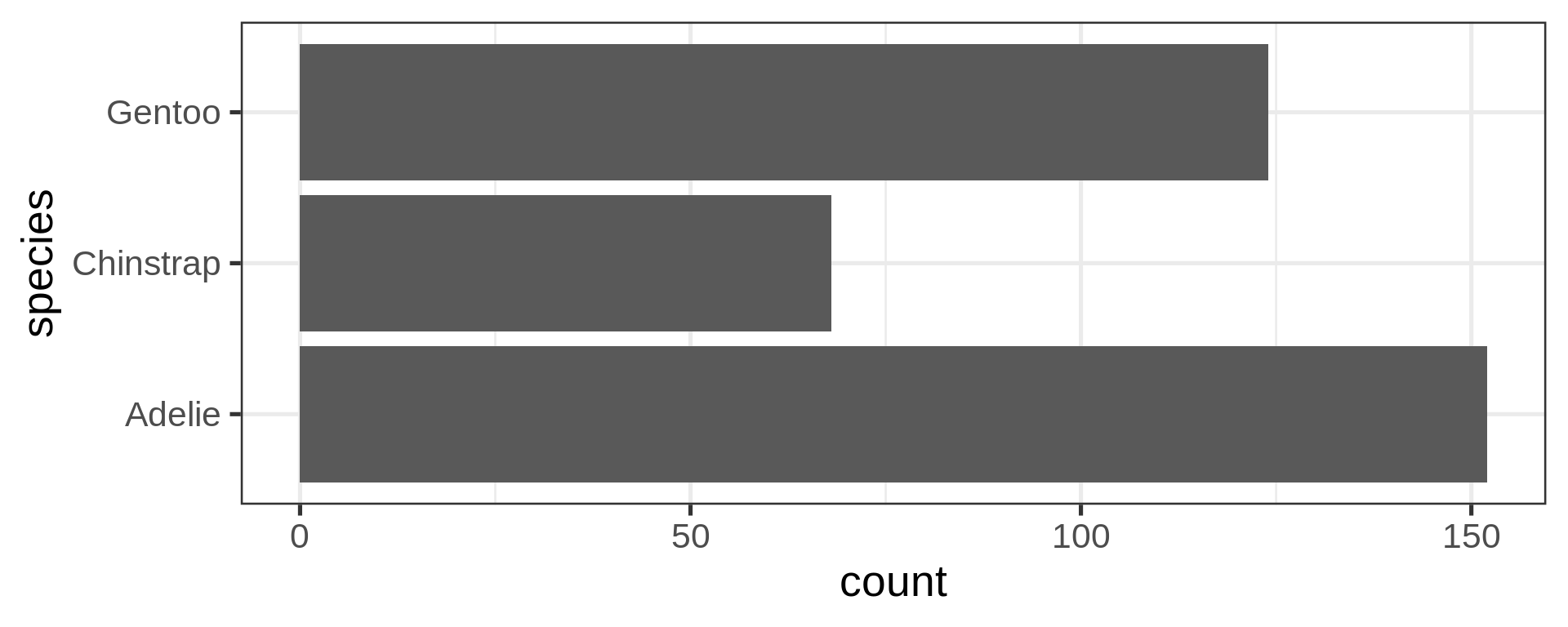
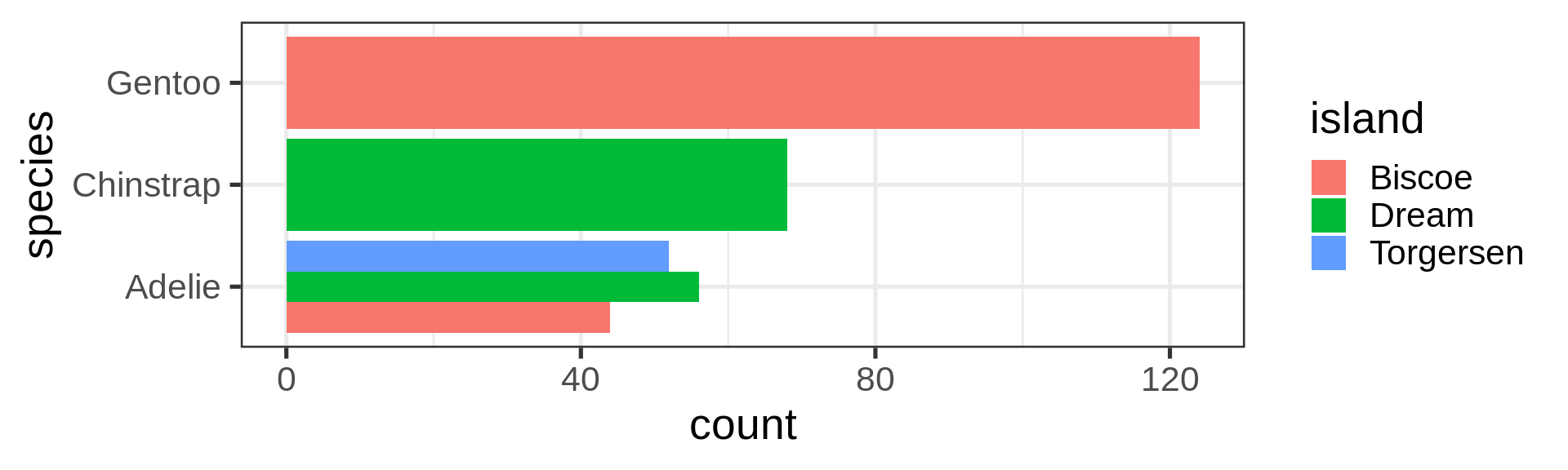
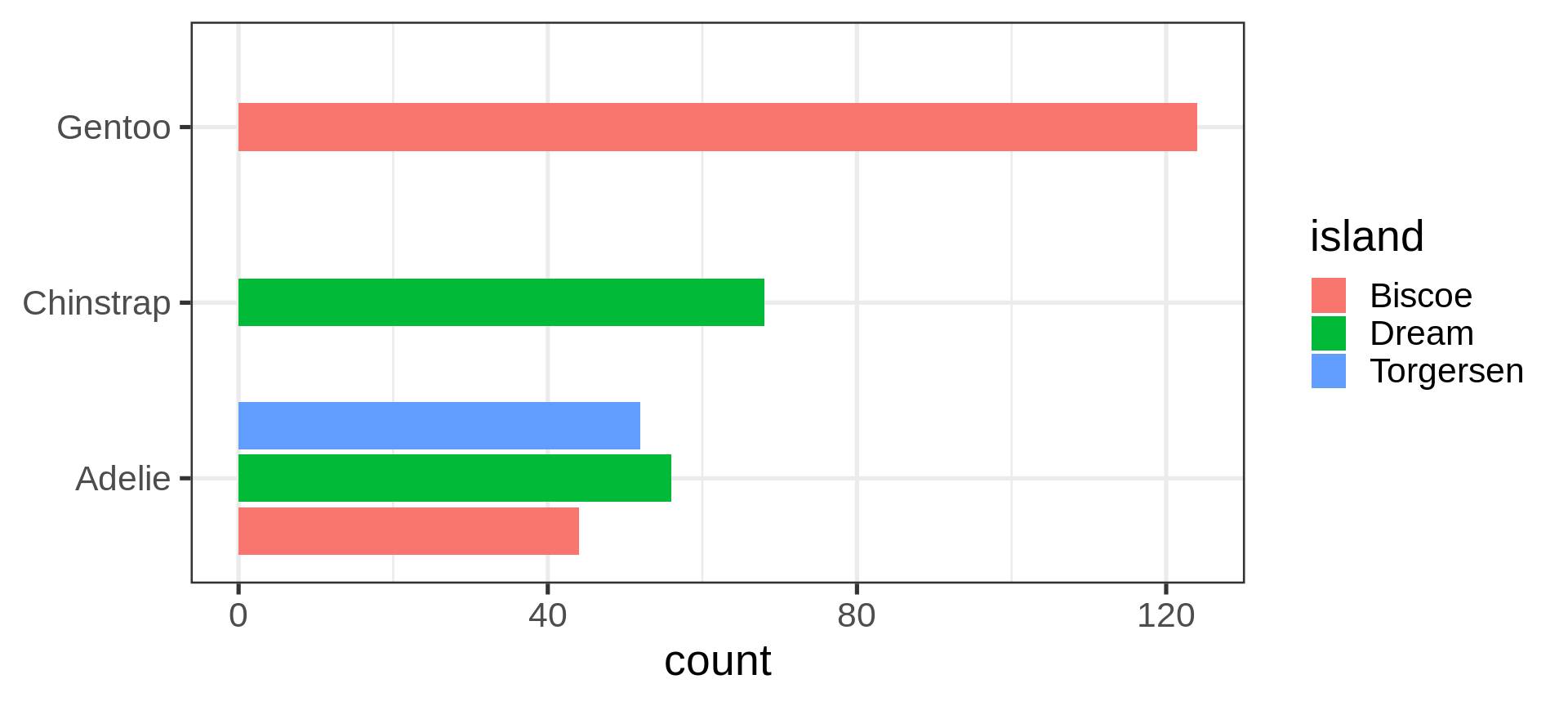
Flexibility in the asthetics for flipping axes
Reorder the categorical variable (forcats)
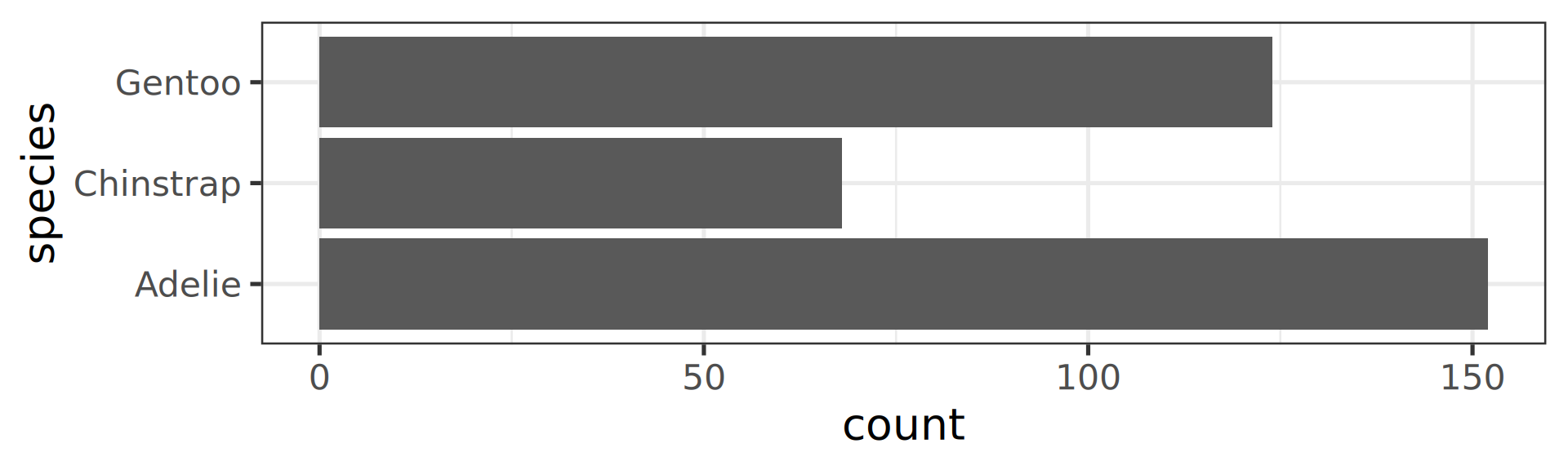
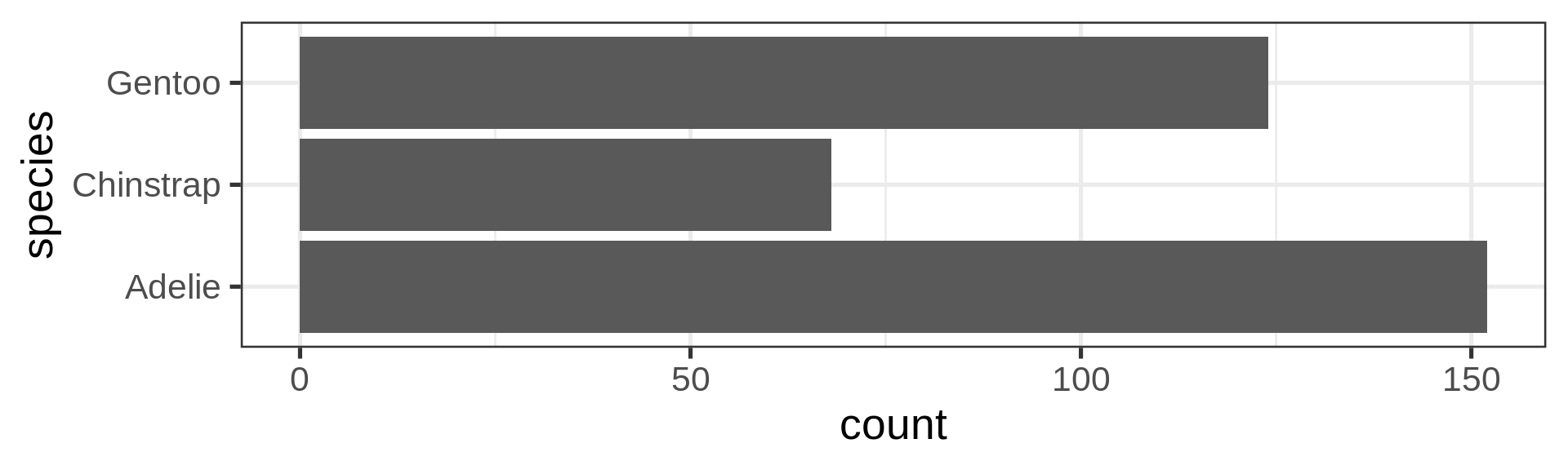
Using the function fct_infreq()
library(forcats)
penguins |>
ggplot(aes(y = fct_infreq(species) |> fct_rev())) +
geom_bar() +
scale_x_continuous(expand = c(0, NA)) +
labs(title = "Palmer penguins species",
y = NULL) +
theme_minimal(14) +
theme(panel.ontop = TRUE,
# better to hide the horizontal grid lines
panel.grid.major.y = element_blank())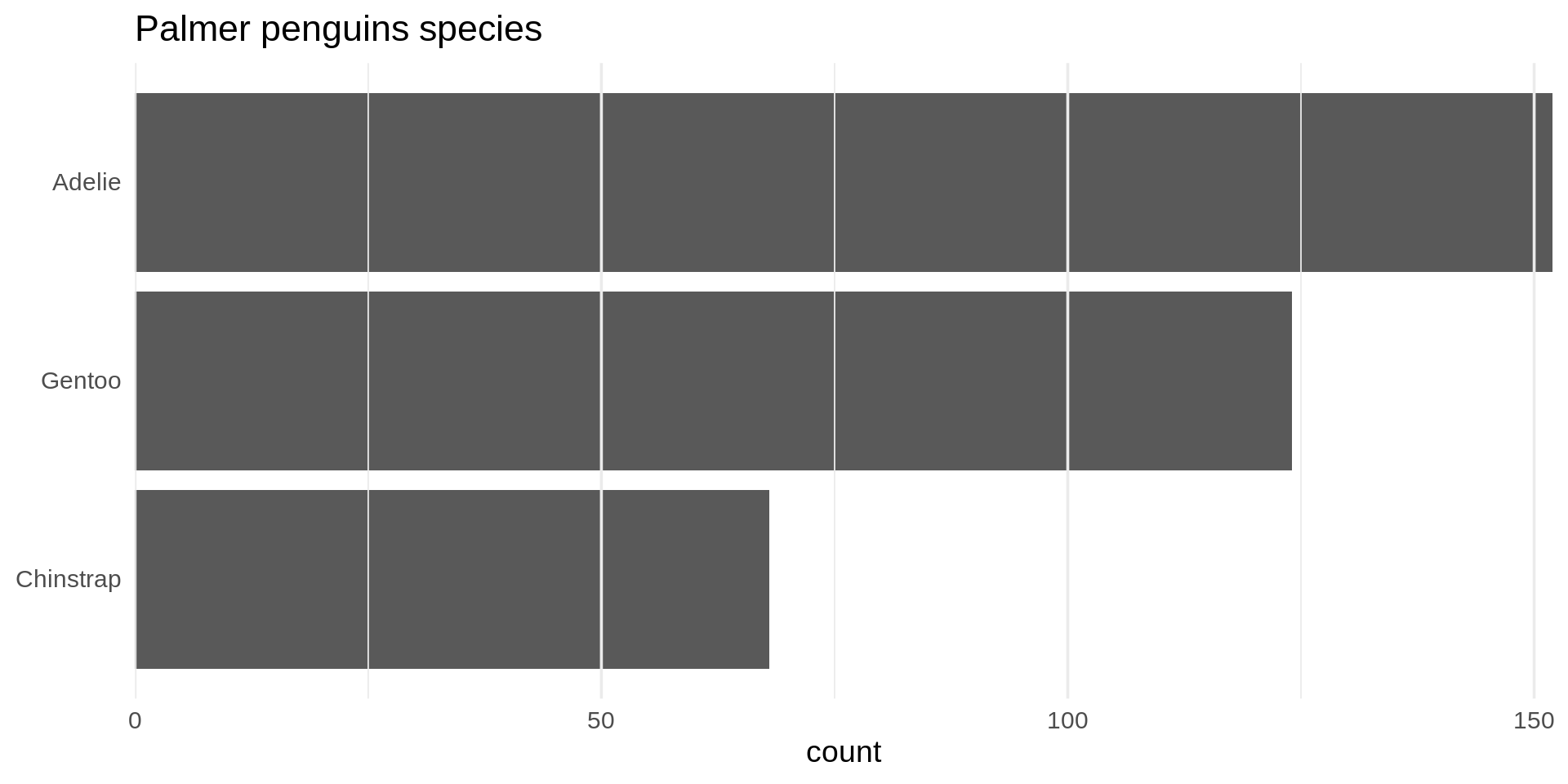
- See the new article FAQ about reordering.
Geometries catalogue
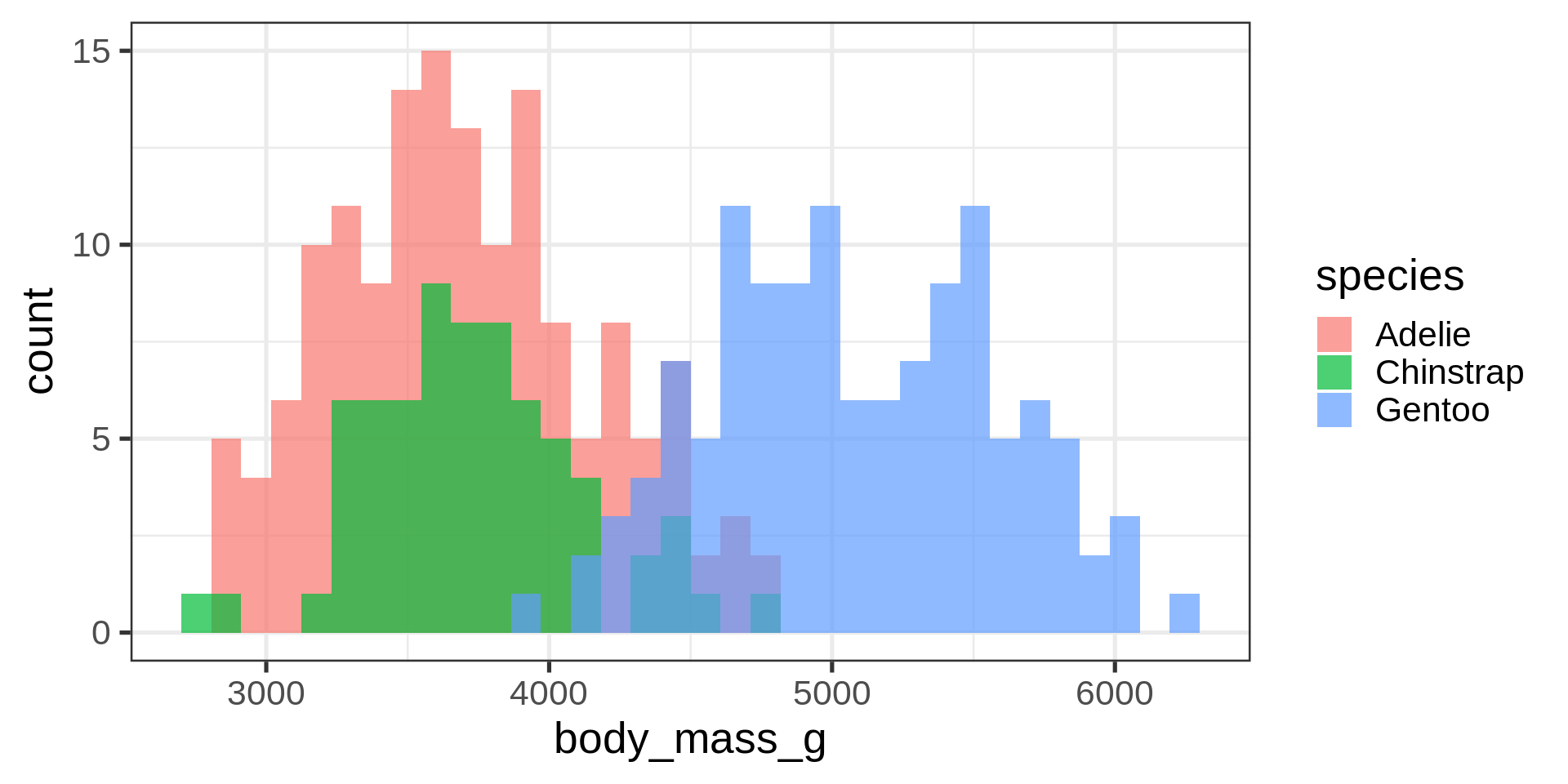
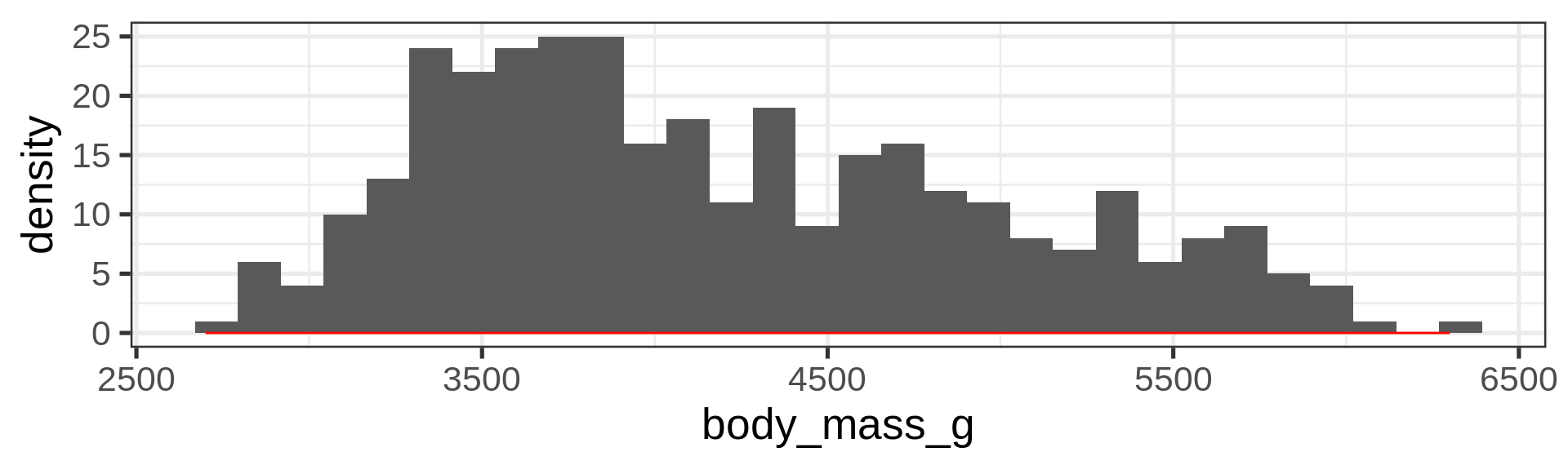
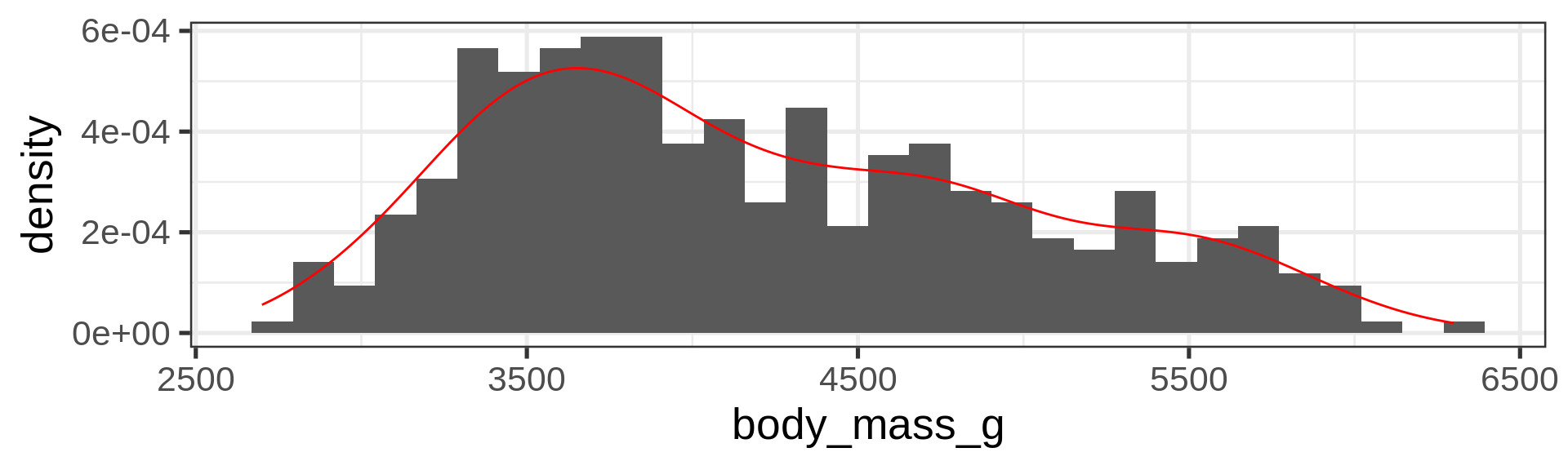
Histograms
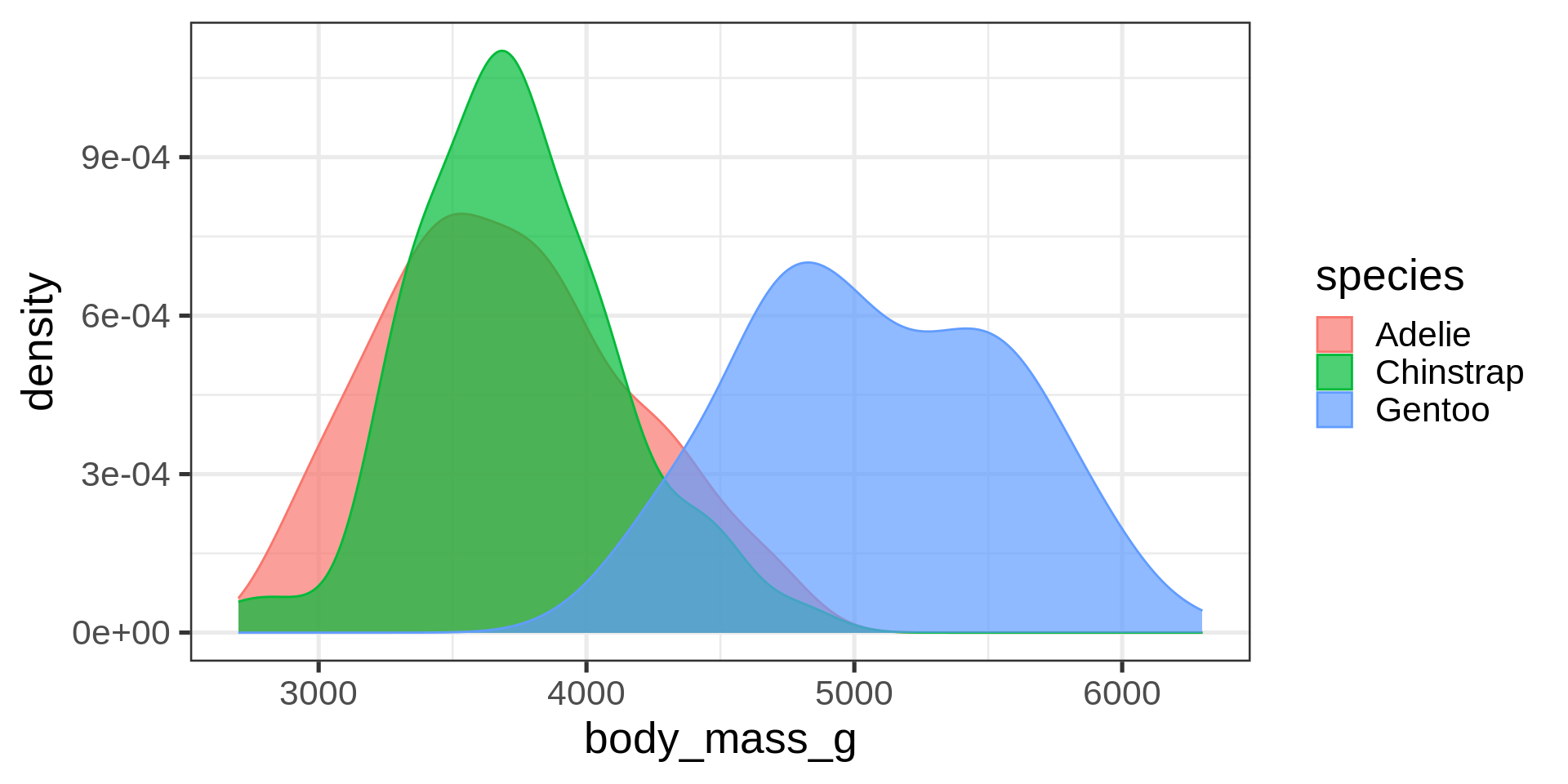
Density plots
Overlay density and histogram
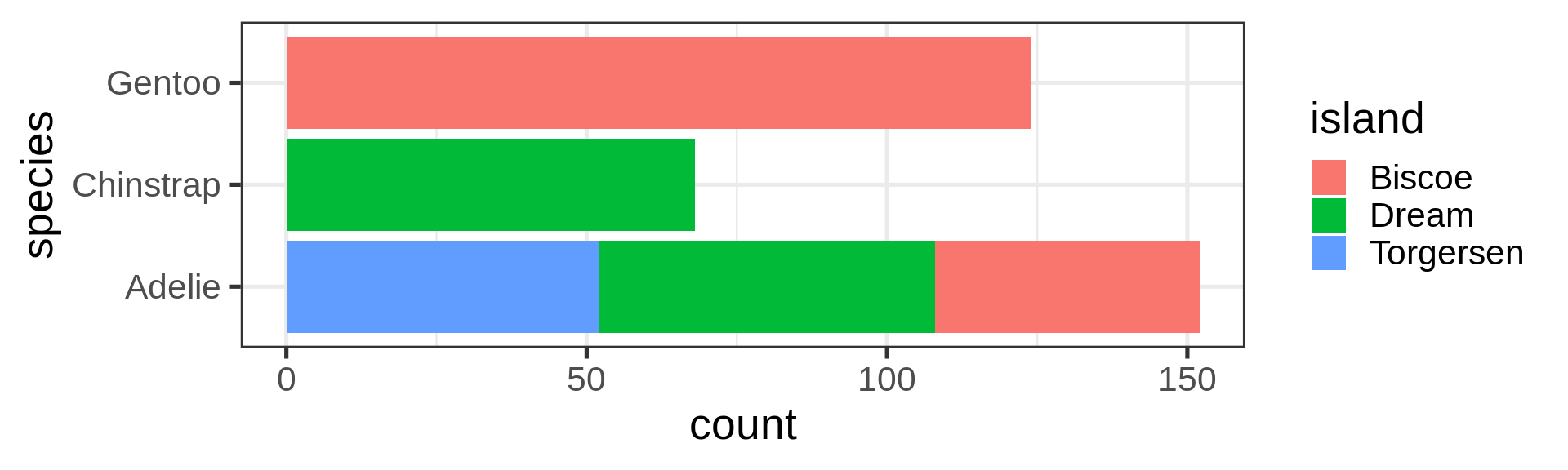
Barplot, categories
Preserve single bar
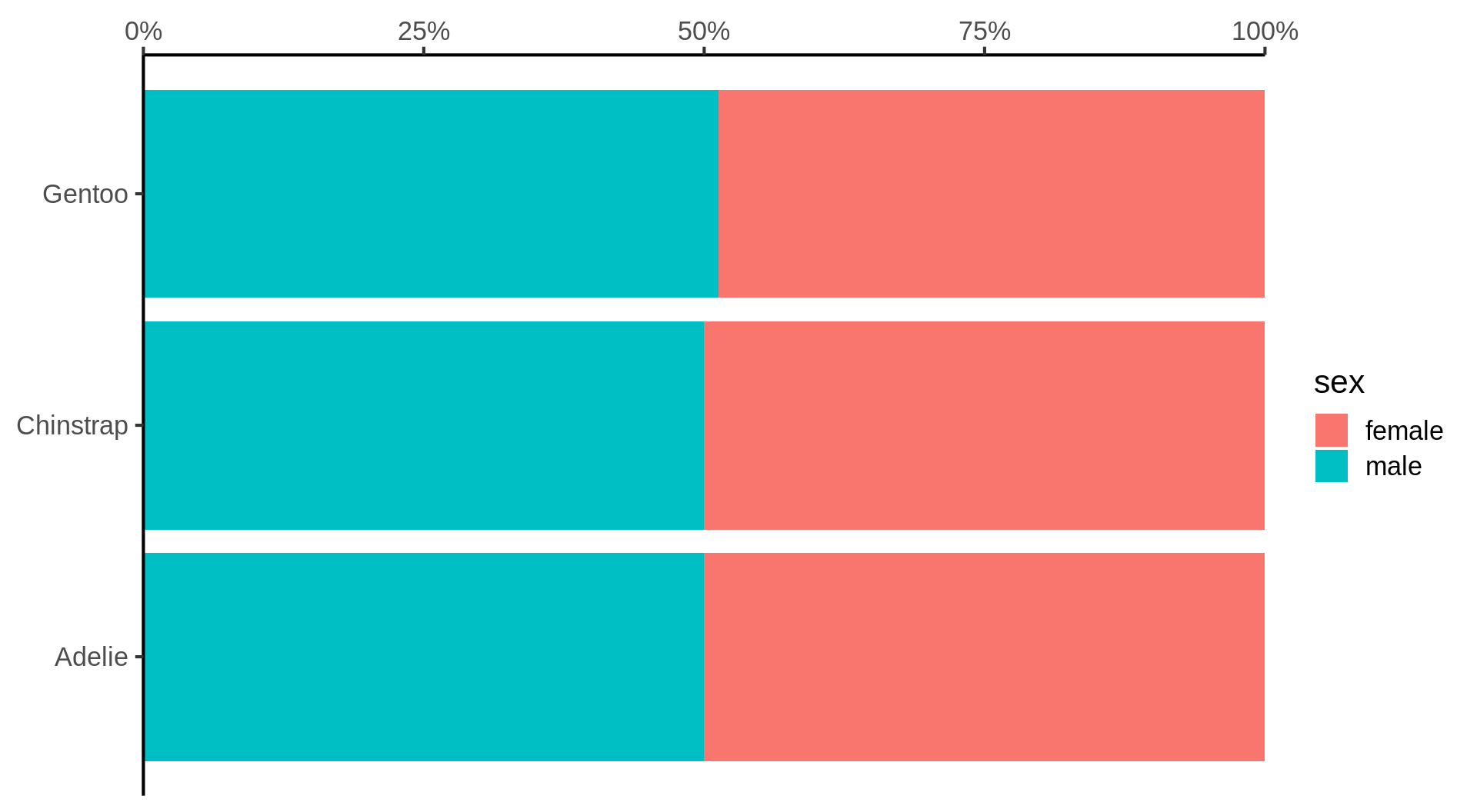
Stacked barchart for proportions
Pie charts involved another coordinate system
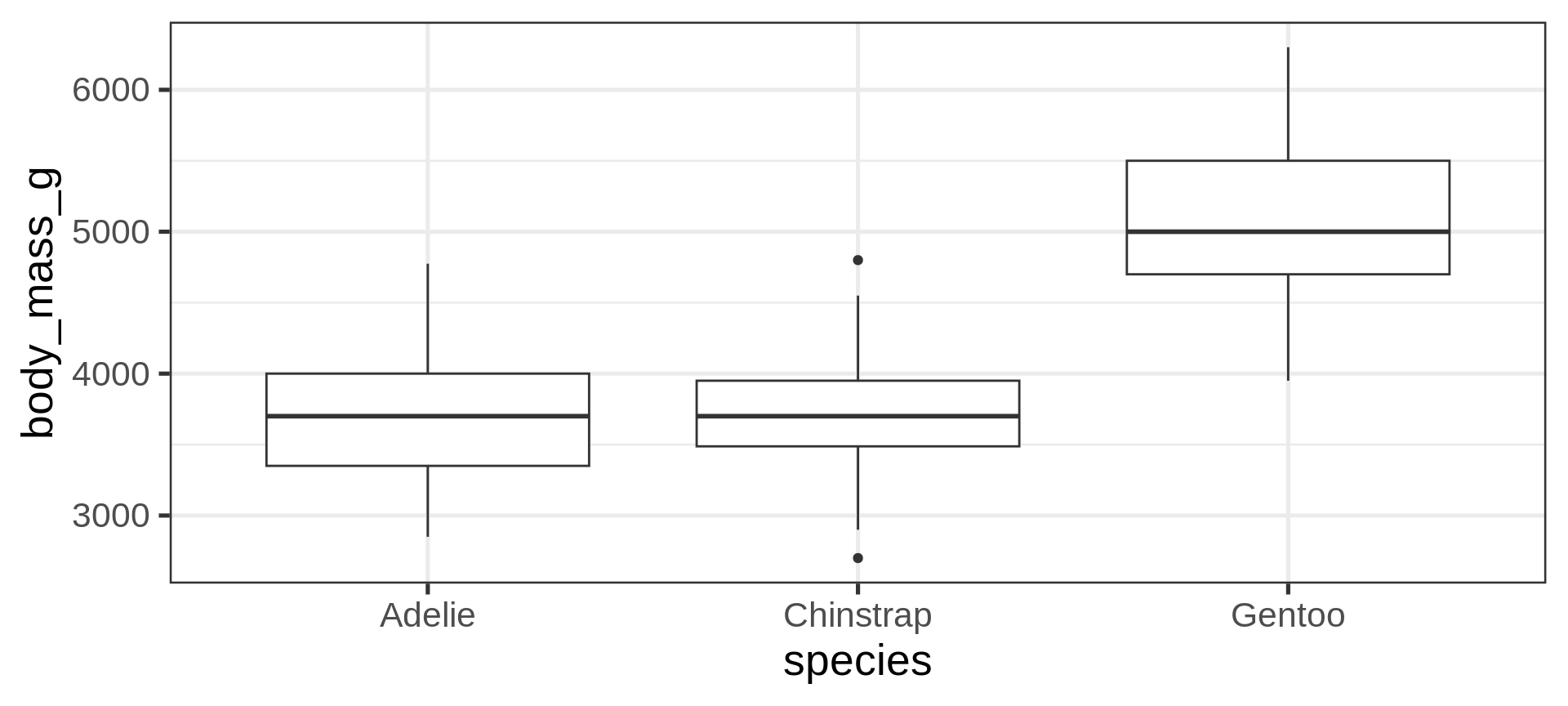
Boxplot, a continuous y by a categorical x
Note
geom_boxplot() is assessing that:
body_mass_gis continuousspeciesis categorical/discrete

Artwork by Allison Horst
Boxplot, dodging by default
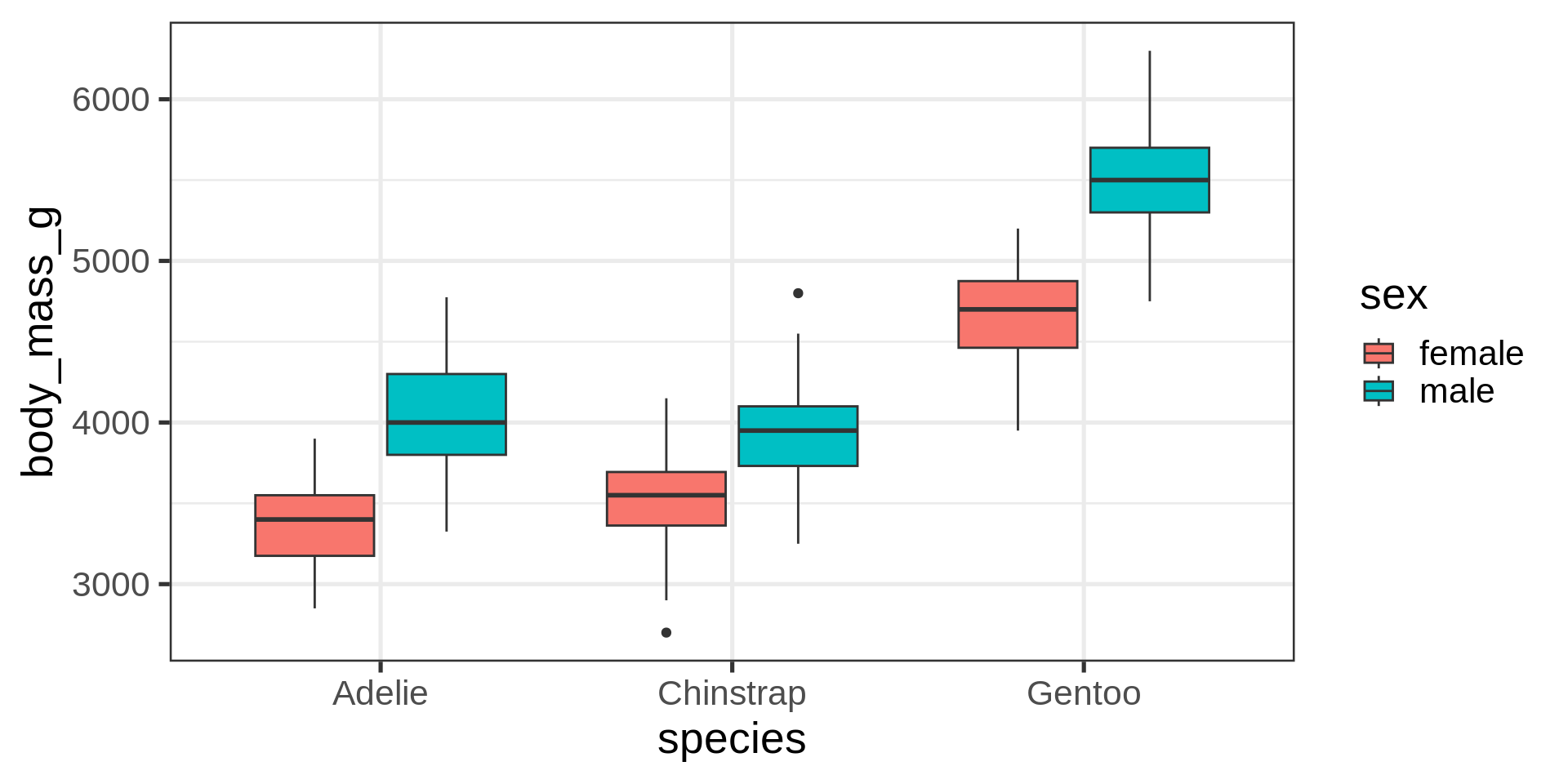
Filter out NA to avoid additional category for missing data.
Better: violin and jitter
Show the data
penguins |>
filter(!is.na(sex)) |>
# define aes here for both geometries
ggplot(aes(y = body_mass_g,
x = species,
fill = sex,
# for violin contours and dots
colour = sex
)) + # very transparent filling
geom_violin(alpha = 0.1, trim = FALSE) +
geom_point(position = position_jitterdodge(dodge.width = 0.9),
# geom_point(position = position_dodge2(width = 0.9 , preserve = "total", padding = 0.005),
alpha = 0.5,
# don't need dots in legend
show.legend = FALSE)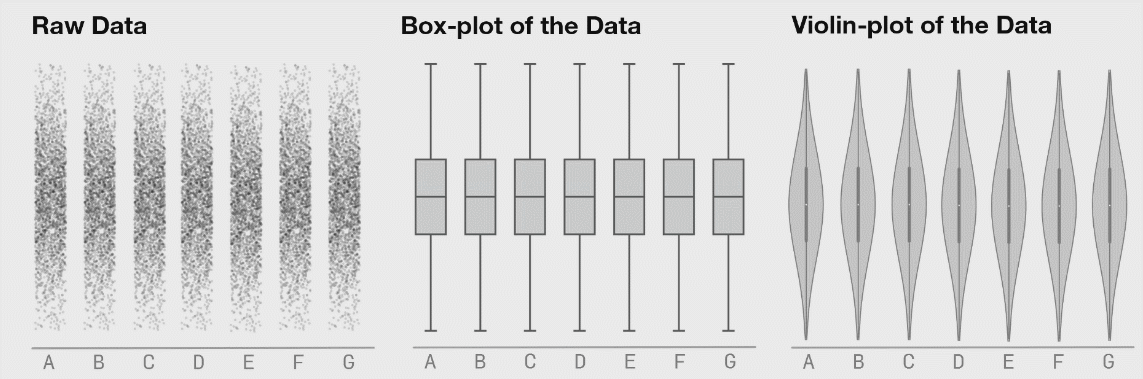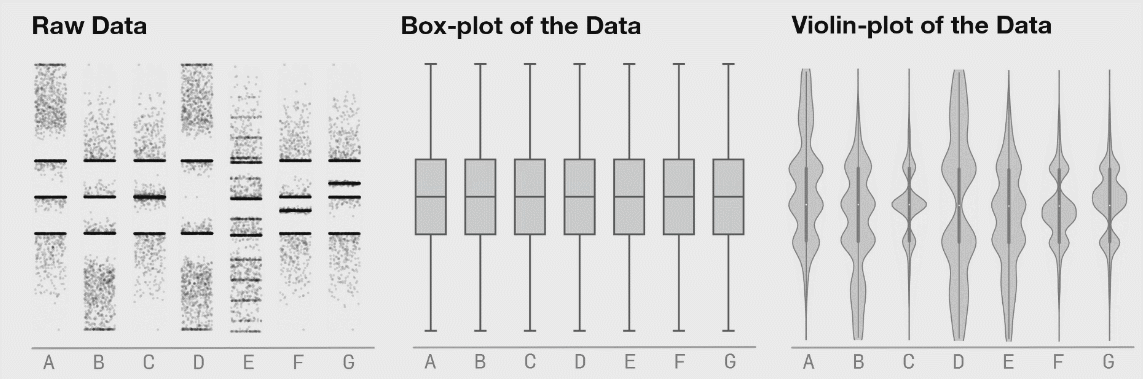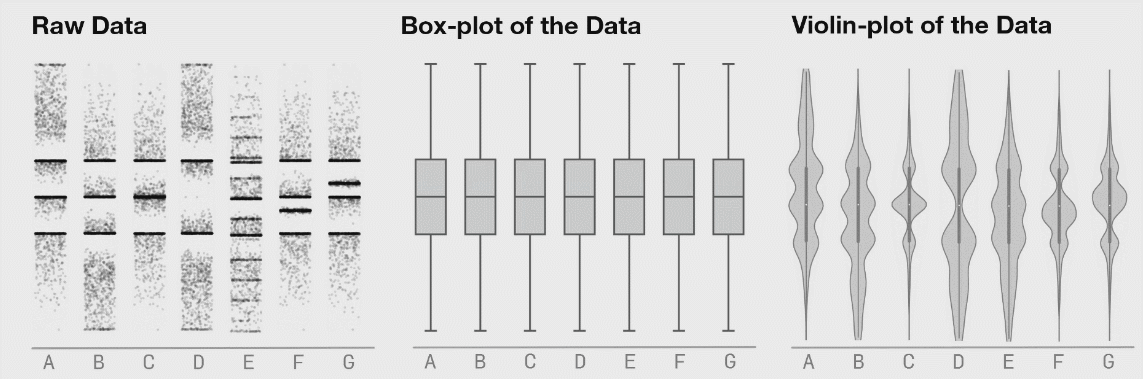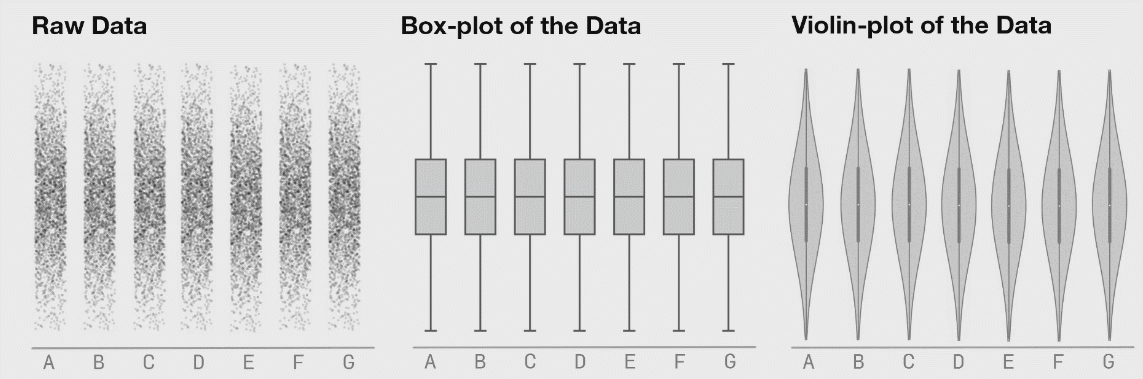
GIF source: Linh Ngo @BioTuring
Coding mistake
What is wrong with this code?
Think about inherited aesthetics!
Error in `geom_smooth()`:
! Problem while computing stat.
ℹ Error occurred in the 2nd layer.
Caused by error in `compute_layer()`:
! `stat_smooth()` requires the following missing aesthetics: x and y.Before we stop
You learned to:
- Apprehend Graphics as a language
- Embrace the layer system
- Link data columns to aesthetics
- Discover geometries
Acknowledgments
- Aurélien Ginolhac
- Thomas Lin Pedersen, great webinar Plotting everything with ggplot2:
- Thinkr (Diane Beldame, Vincent Guyader, Colin Fay)
- Allison M. Horst, Alison P. Hill and Kristen B. Gorman
* - Romain Lesur (
pagedown), Christophe Dervieux - Hadley Wickham
Further reading
- ggplot2 book by H. Wickham (free access)
- Official FAQ, axes, facets, custom, annot, reorder, bars
- R for Data Science by H. Wickham / G. Gromelund (free access)
- TidyTuesday challenges
- Vizualisation gallery by Cédric Scherer
- plotting tutorial by Cédric Scherer