import matplotlib.pyplot as plt
# And while we're at it - for data input
import plotly.express as pxData visualisation in Python
Matplot, Seaborn and Plotly
MADS6
2026-03-04
Introduction
“By visualizing information, we turn it into a landscape that you can explore with your eyes, a sort of information map. And when you’re lost in information, an information map is kind of useful.” – David McCandless
Data visualization
Learning objectives
- A look at relevant plotting libraries
- From
matplotlibtoseaborn - Using
seabornfor- Categorical, relational and distributional exploration
- Interactive graphs with
plotly’s main chart types
Materials
Data visualisation in Python
What story is behind your data?
Why was this data collected, and how?
Is your data collected to find trends?
To compare different options?
Is it showing some distribution?
Or is used to observe the relationship between different value sets?
Know the source of your data!
Understanding the origin story of your data and knowing what it’s trying to deliver will make choosing a chart type and a library a much easier task for you.
Aims for visualising data
Exploration
Explore a novel data set visually
Column or variable names are legit labels
Themes minimal
Possible questions
Are outliers interesting data points or noise?
Can we find correlation?
Are there unusual distributions of data?
Do we need to transform?
Audience is user
Communication
At least structure of the data set is known
Axis labels are easily understandable
Title and caption complement the graph
Graph has a story to tell
Themes support message
Audience are a larger group who might not know the data
Python has many plotting libraries

Matplotlib

Probably the best known library for Python, started to be developed in 2003.
Aims to emulate the commands of the
MATLABsoftware, which was the scientific standard back then.Several features, such as the global style of MATLAB, were introduced to make the transition to
matplotlibeasier for MATLAB users.
matplotlib.pyplot submodule
For most of our plotting tasks the
pyplotmodule provides a functional plotting interface.Rather than importing the whole matplotlib package, we will only import the
pyplotmodule using the dot (.) notation.pyplotcontains a simpler interface – plot the data without explicitly configuring the Figure and Axes themselves.
A first plot
Basic plotting
- Lists as input
- No axis labels
- Shorthand codes for labels
Structure of plots in Matplotlib
All matplotlib objects are inherited from the Artist abstract base class.
Each plot is encapsulated in a Figure object.
The Figure is the top-level container of the visualization.
It can have multiple Axes, which are basically individual plots inside this top-level container.
Python objects control axes, tick marks, legends, titles, text boxes, the grid, and many other objects.
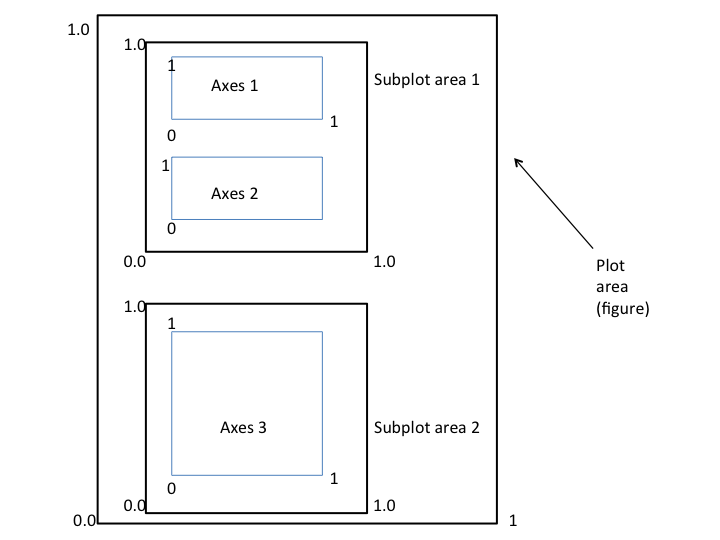
Figure structure
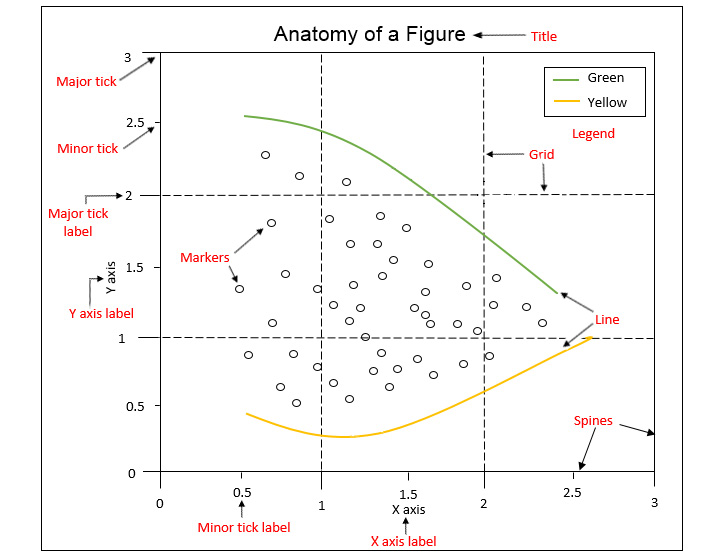
Figure manipulation
Have to be displayed with the command
plt.show().Figures that are no longer used should be closed by explicitly calling
plt.close().To save a figure you can use the command
plt.savefig("fname").

“Gapminder identifies systematic misconceptions about important global trends and proportions and uses reliable data to develop easy to understand teaching materials to rid people of their misconceptions.”
| country | continent | year | lifeExp | pop | gdpPercap | iso_alpha | iso_num | |
|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | Asia | 1952 | 28.801 | 8425333 | 779.445314 | AFG | 4 |
| 1 | Afghanistan | Asia | 1957 | 30.332 | 9240934 | 820.853030 | AFG | 4 |
| 2 | Afghanistan | Asia | 1962 | 31.997 | 10267083 | 853.100710 | AFG | 4 |
| 3 | Afghanistan | Asia | 1967 | 34.020 | 11537966 | 836.197138 | AFG | 4 |
| 4 | Afghanistan | Asia | 1972 | 36.088 | 13079460 | 739.981106 | AFG | 4 |
Let’s explore just four basic chart types.
Bar Chart
Text(0.5, 1.0, 'Life expectancy by continent in 2002')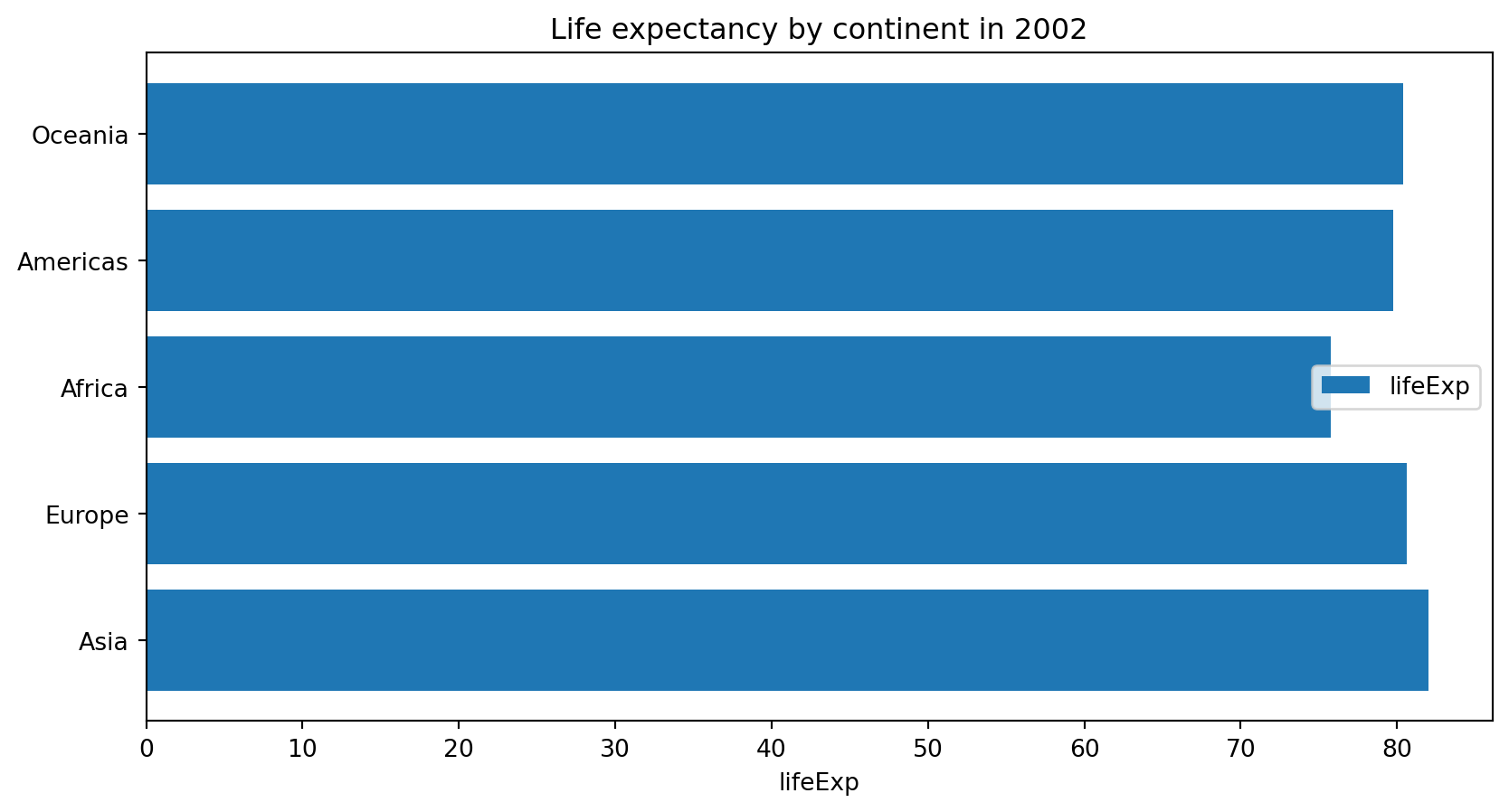
plt.bar(x, height, [width])- vertical bar plot.plt.barh()- horizontal bar plot
Anything wrong about this plot?
Line plot
for country in ["Nigeria", "Belgium",
"China", "Kuwait"]:
plt.plot(
gapminder[gapminder["country"] ==
country]["year"],
gapminder[gapminder["country"] ==
country]["gdpPercap"],
label = country)
plt.yscale('log')
plt.title("GDP per cap over the years", fontsize=24)
plt.ylabel('gdpPercap', fontsize=20)
plt.legend()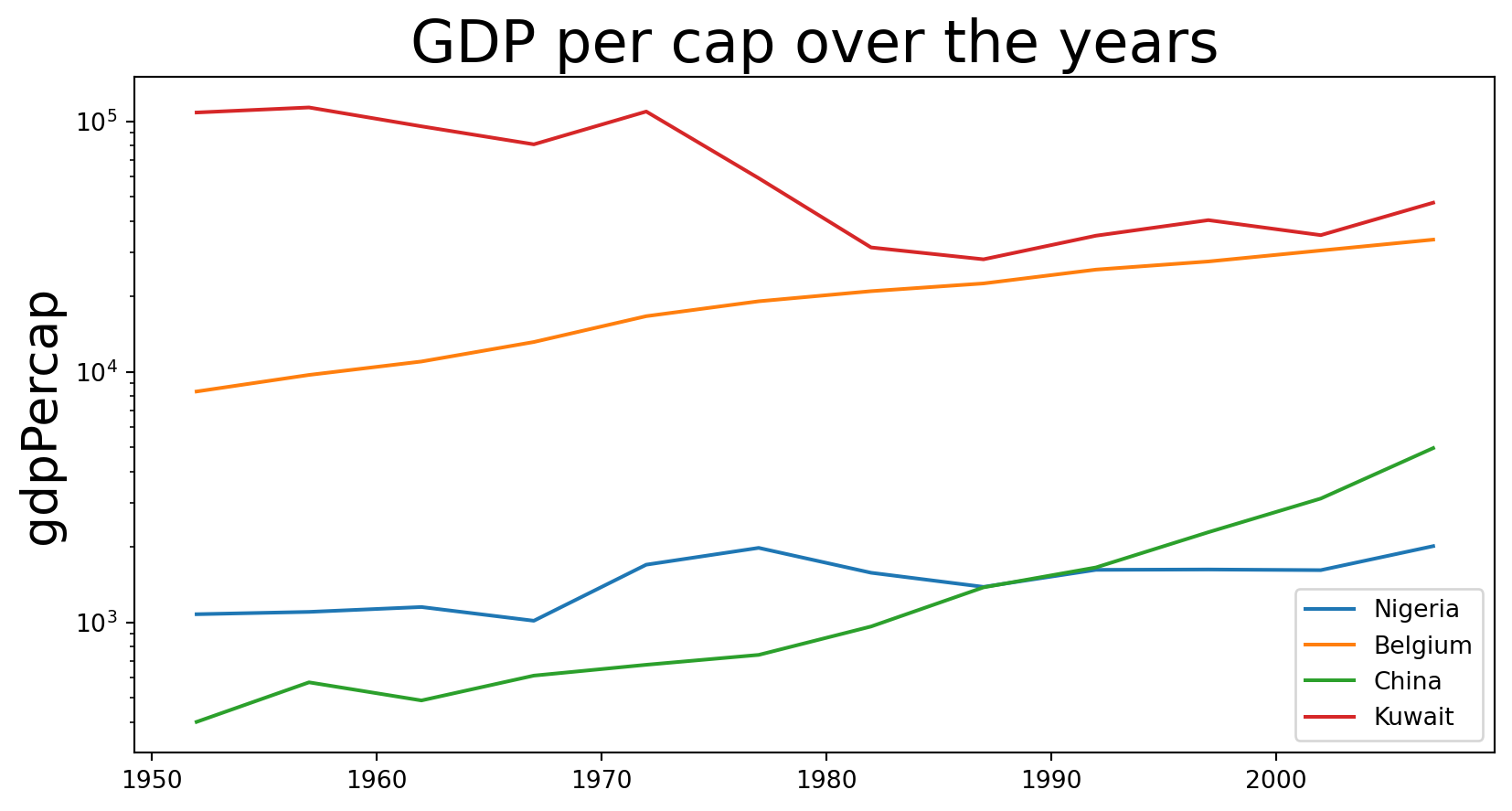
Legend place by algorithm
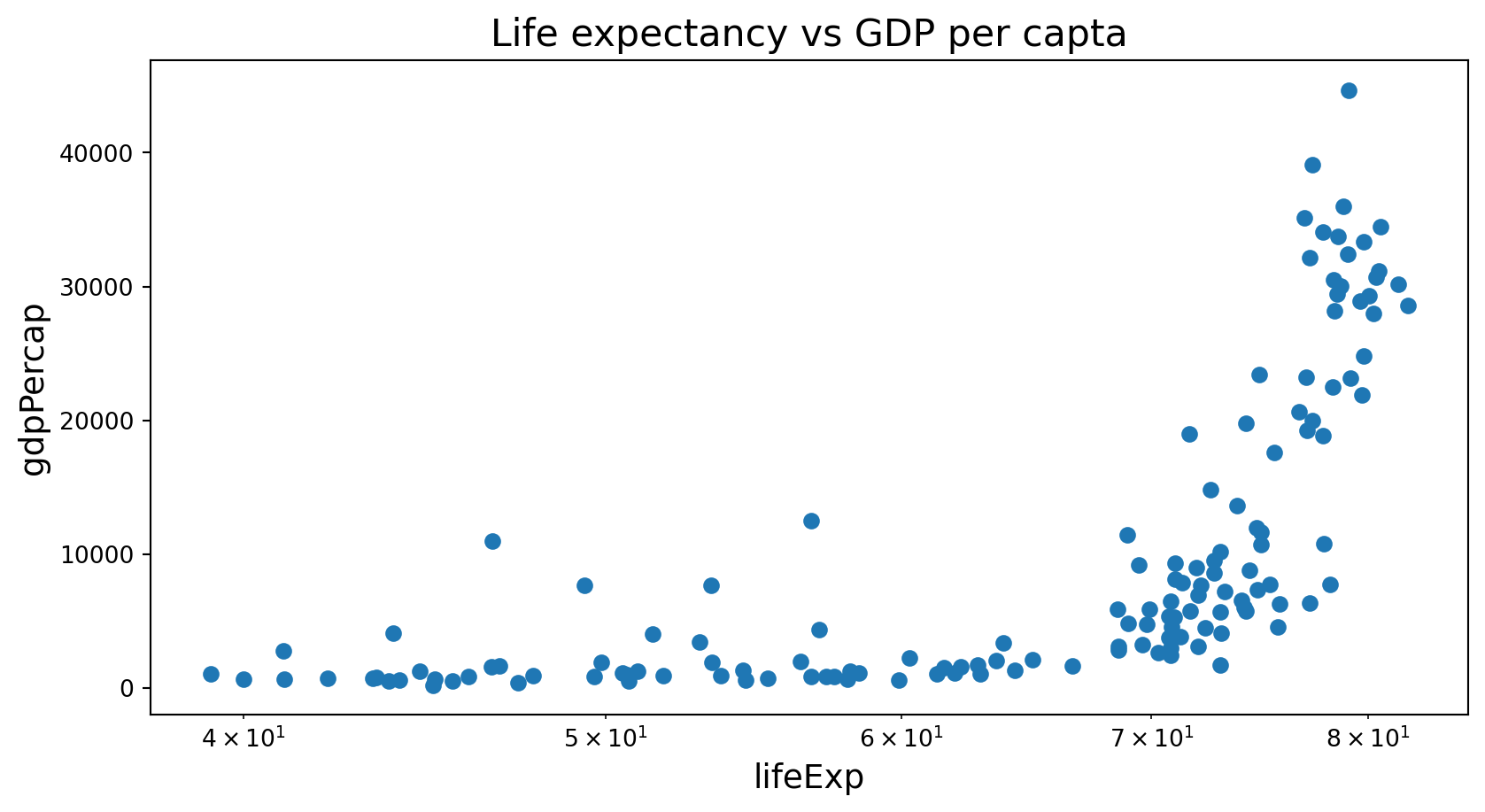
Scatter plot
Histogram
Text(0.5, 0, 'lifeExp')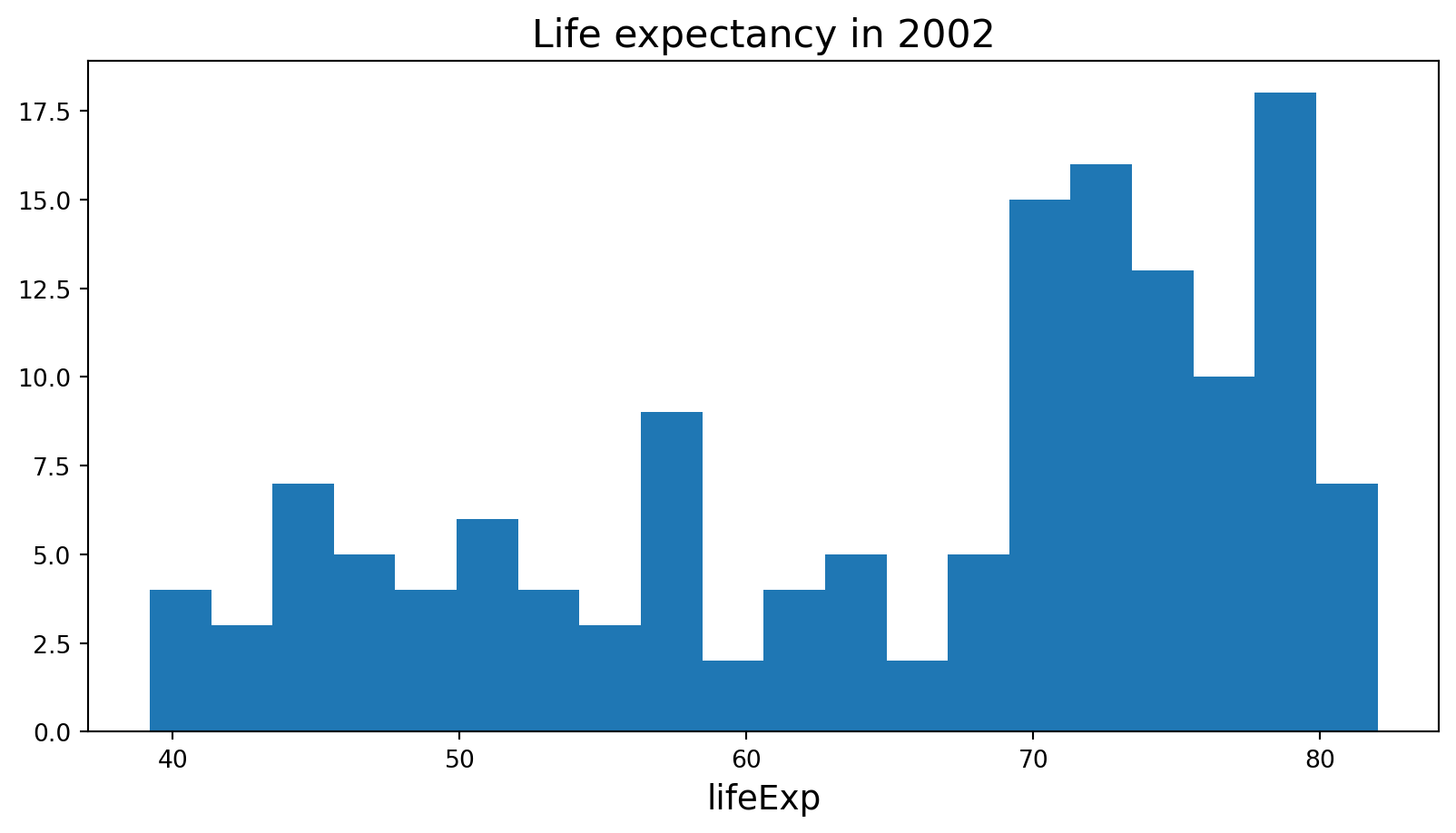
Histogram
A histogram is a bar plot where the axis representing the data variable is divided into a set of discrete bins.
The count of observations falling within each bin is shown using the height of the corresponding bar.

seabornis a Python data visualization library based onmatplotlib. It provides a high-level interface for drawing attractive and informative statistical graphics.It builds on top of
matplotliband integrates closely with pandas data structures.Less adjustments have to be done than in
matplotlib.
“If
matplotlib“tries to make easy things easy and hard things possible”,seaborntries to make a well-defined set of hard things easy to do.”
Seaborn
Advantages
No additional data wrangling to be able to plot the data from the DataFrames as in Matplotlib.
Operate on DataFrames and full dataset arrays.
Internally performs the necessary semantic mappings and statistical aggregation to produce informative plots.
Beautiful out-of-the-box plots with different themes.
Built-in color palettes that can be used to reveal patterns in the dataset.
A high-level abstraction that still allows for complex visualizations.
Disadvantages
- Matplotlib dependency
Overview of seaborn plotting functions
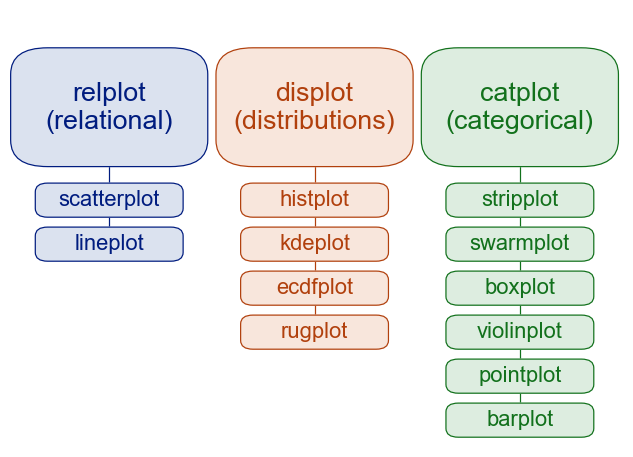
Figure-level vs. axes-level functions
In addition to the module classification, seaborn functions are sub-classified as:
- axes-level plot data onto a single
matplotlib.pyplot.Axesobject - figure-level interface with
matplotlibthrough aseabornobject that manages the figure.
Setup for Seaborn usage
Palmer penguins
The data is available in GitHub. The goal is to provide a great data set for data exploration & visualization, as an alternative to the iris data set.

Load the data
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
Distribution plots
Exploration
Analyze or model data to understand how the variables are distributed.
Techniques for distribution visualization can provide quick answers.
Good questions
- What range do the observations cover?
- What is their central tendency?
- Are they heavily skewed in one direction?
- Is there evidence for bimodality? Are there significant outliers?
- Do the answers to these questions vary across subsets defined by other variables?
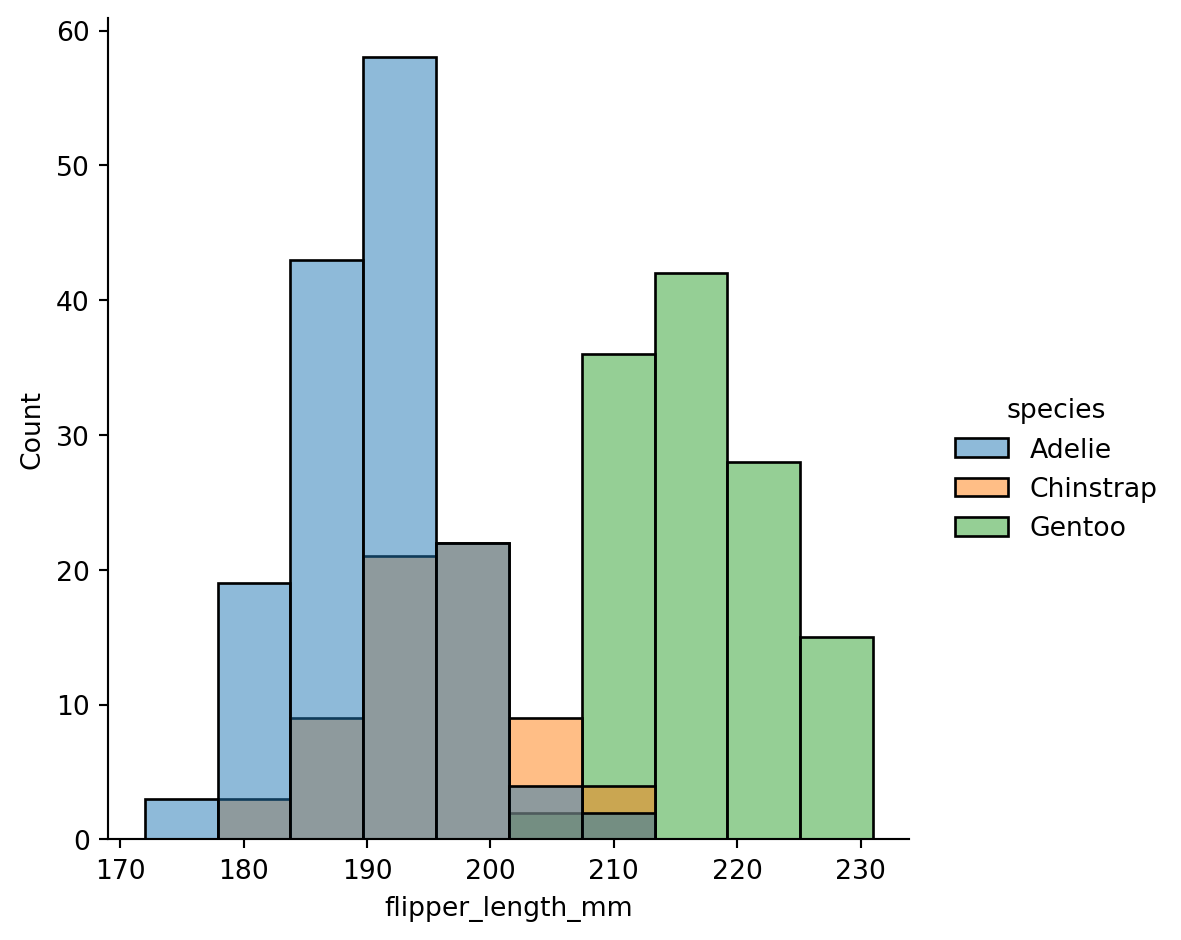
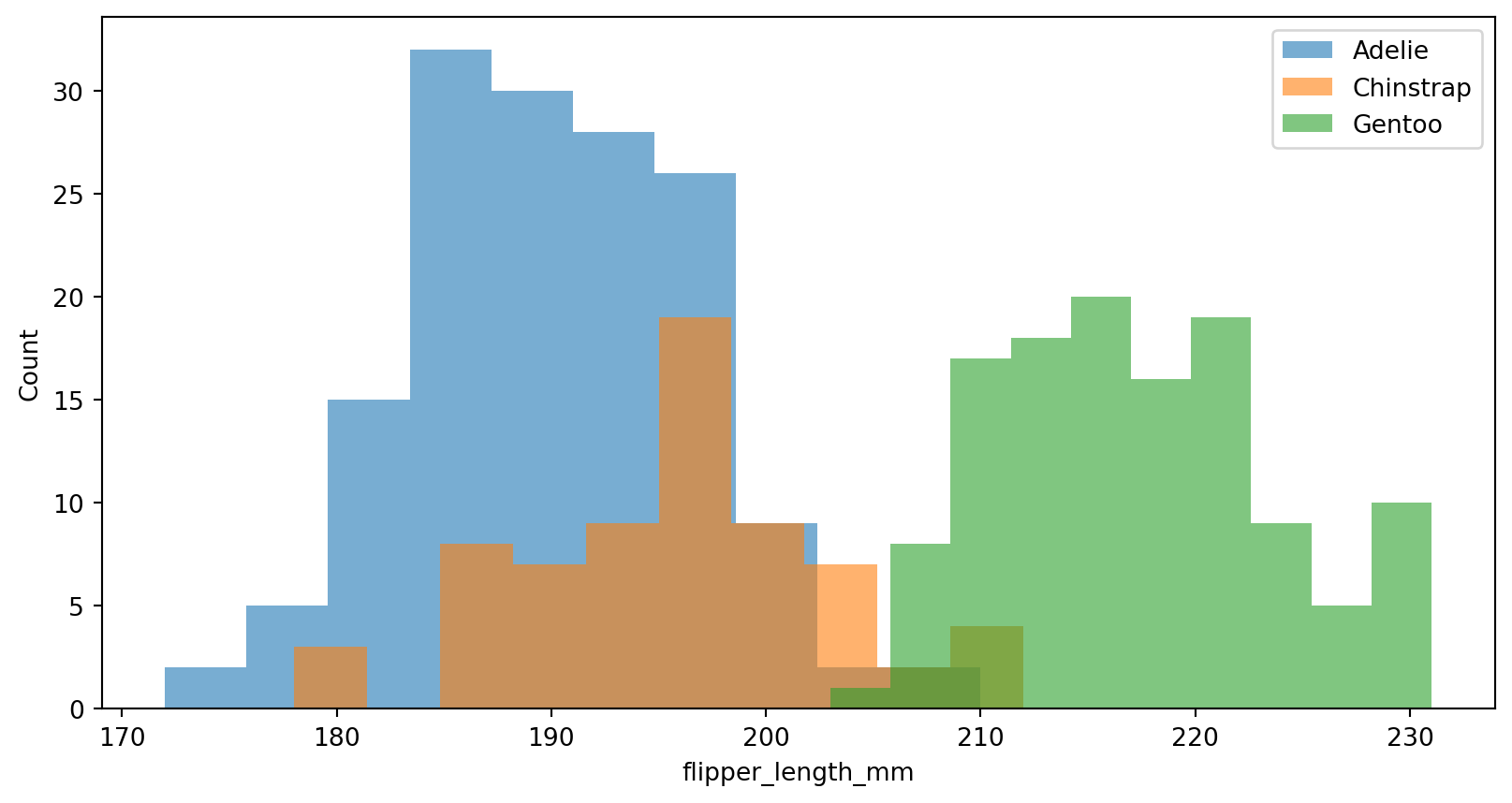
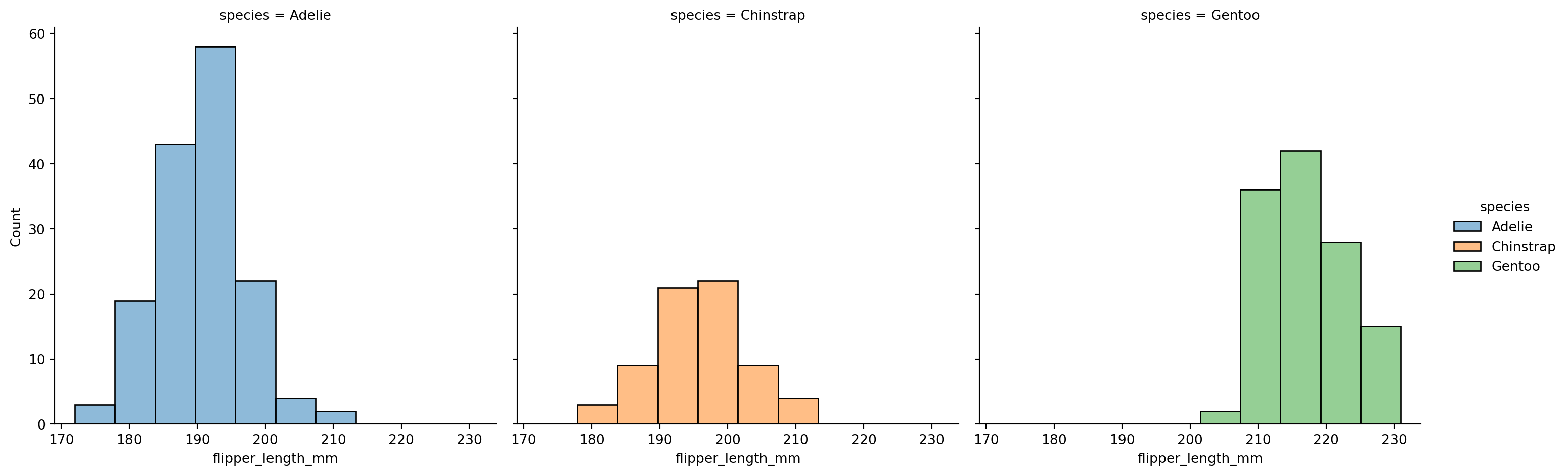
Histograms - histplot()
How would look like in matplotlib?
How would this look like in ggplot2?
#| eval: false
library(ggplot2)
library(palmerpenguins)
ggplot(penguins,
aes(x = flipper_length_mm,
fill = species)) +
geom_histogram(
color = "#e9ecef",
alpha = 0.6,
position = 'identity',
bins = 10
) +
theme(legend.position = c(0.87, 0.75))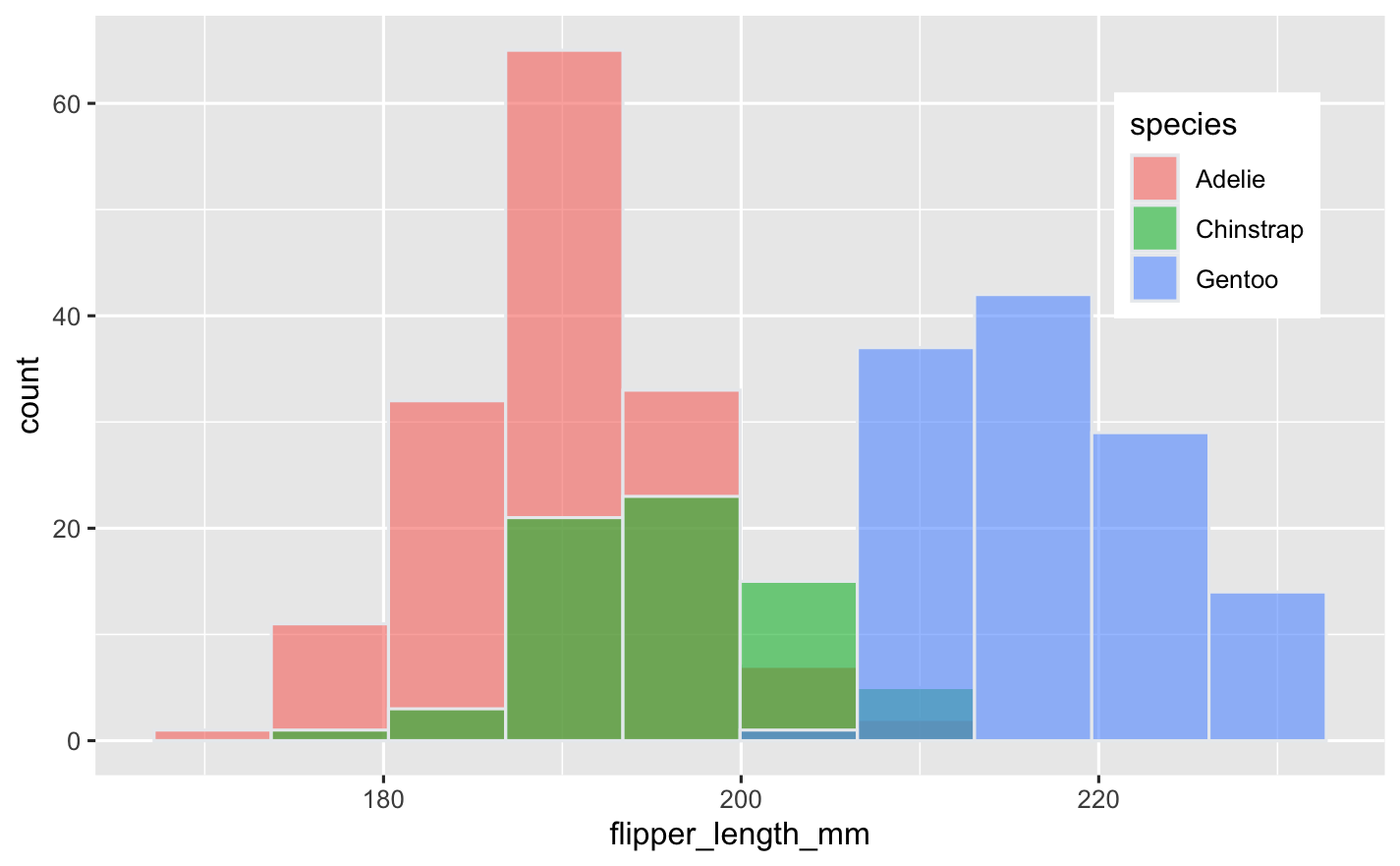
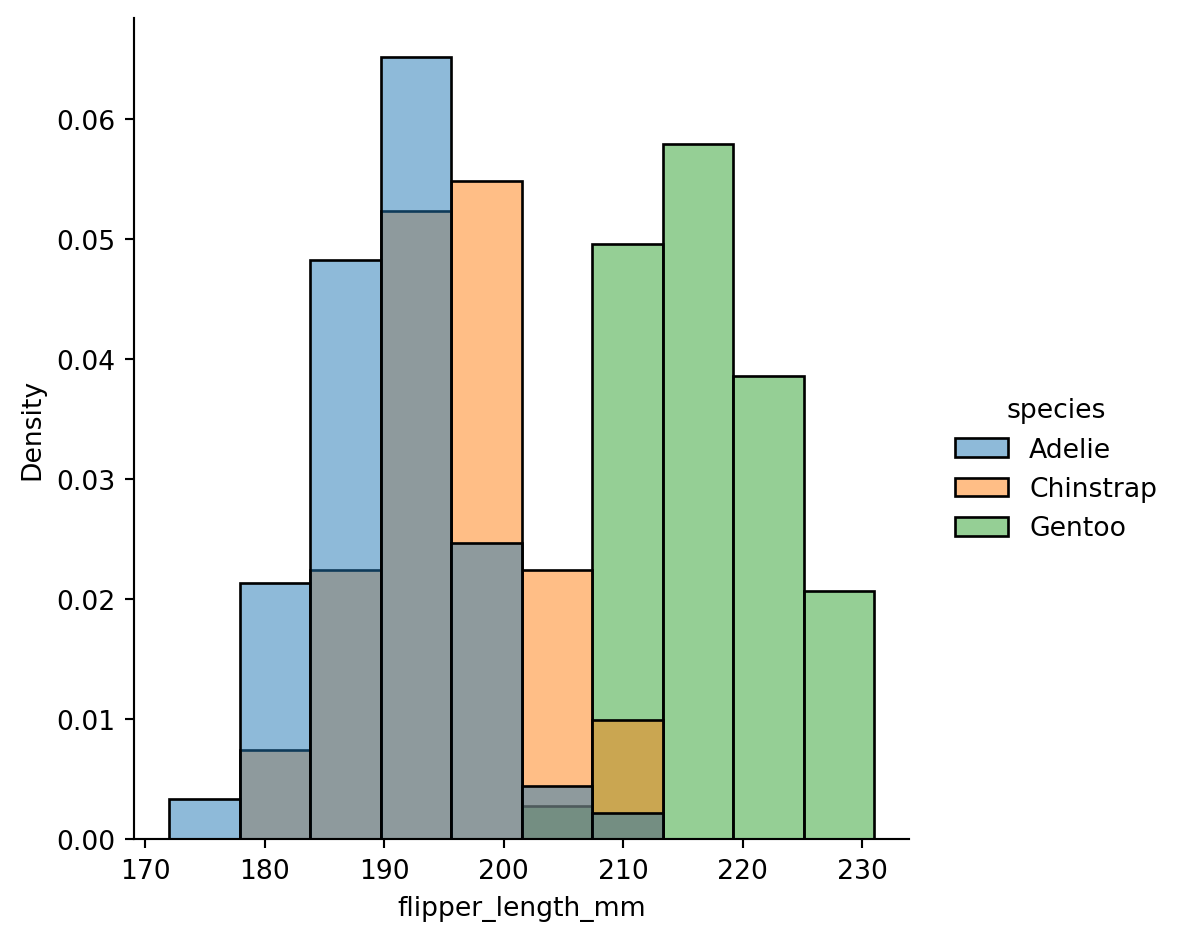
Normalized histogram statistics
When the subsets have unequal numbers of observations, comparing their distributions in terms of counts may not be ideal.
One solution is to normalize the counts using the
statparameter:
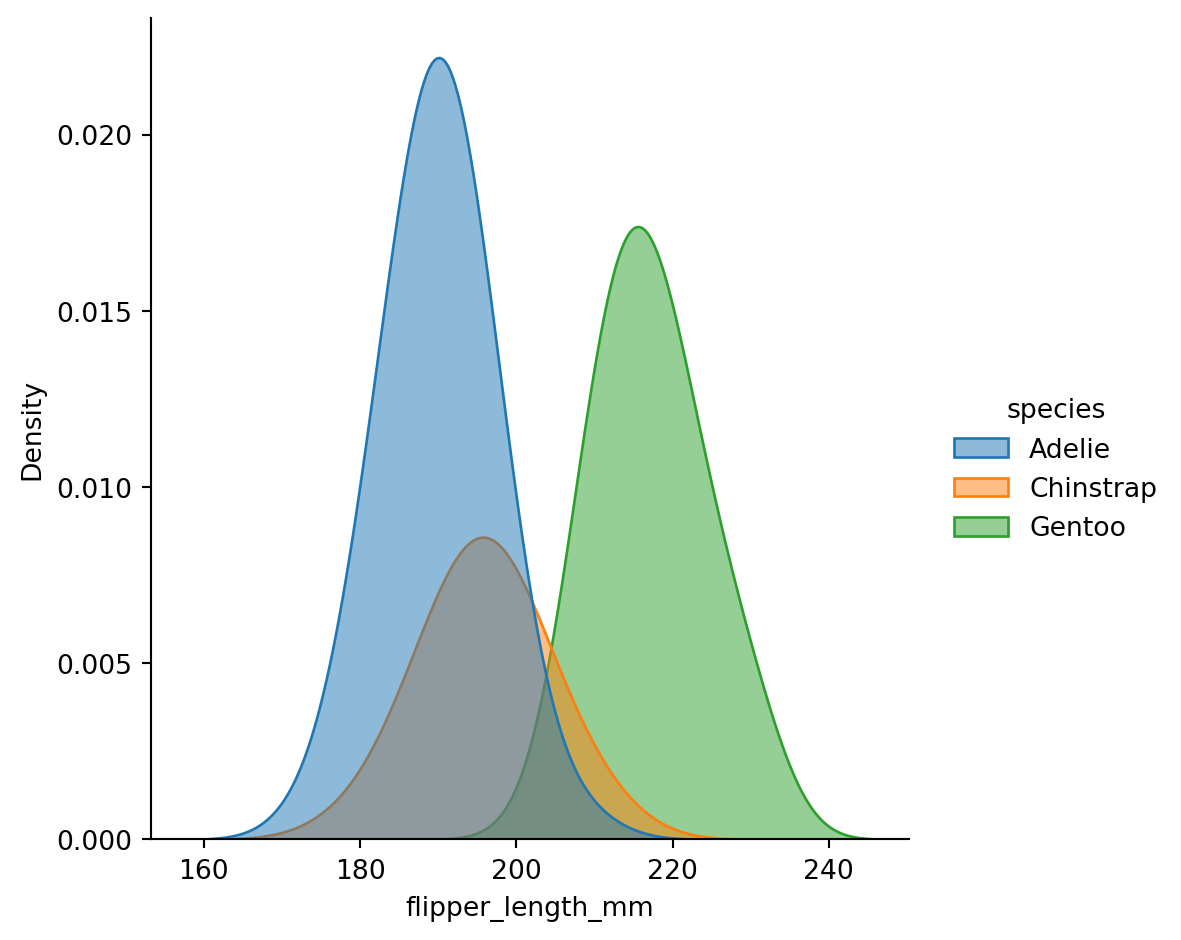
Kernel - kdeplot()
A histogram approximates the underlying probability density function that generated the data by binning and counting observations.
Kernel density estimation (KDE) presents a different solution to the same problem.
A KDE plot smooths the observations with a Gaussian kernel, producing a continuous density estimate.
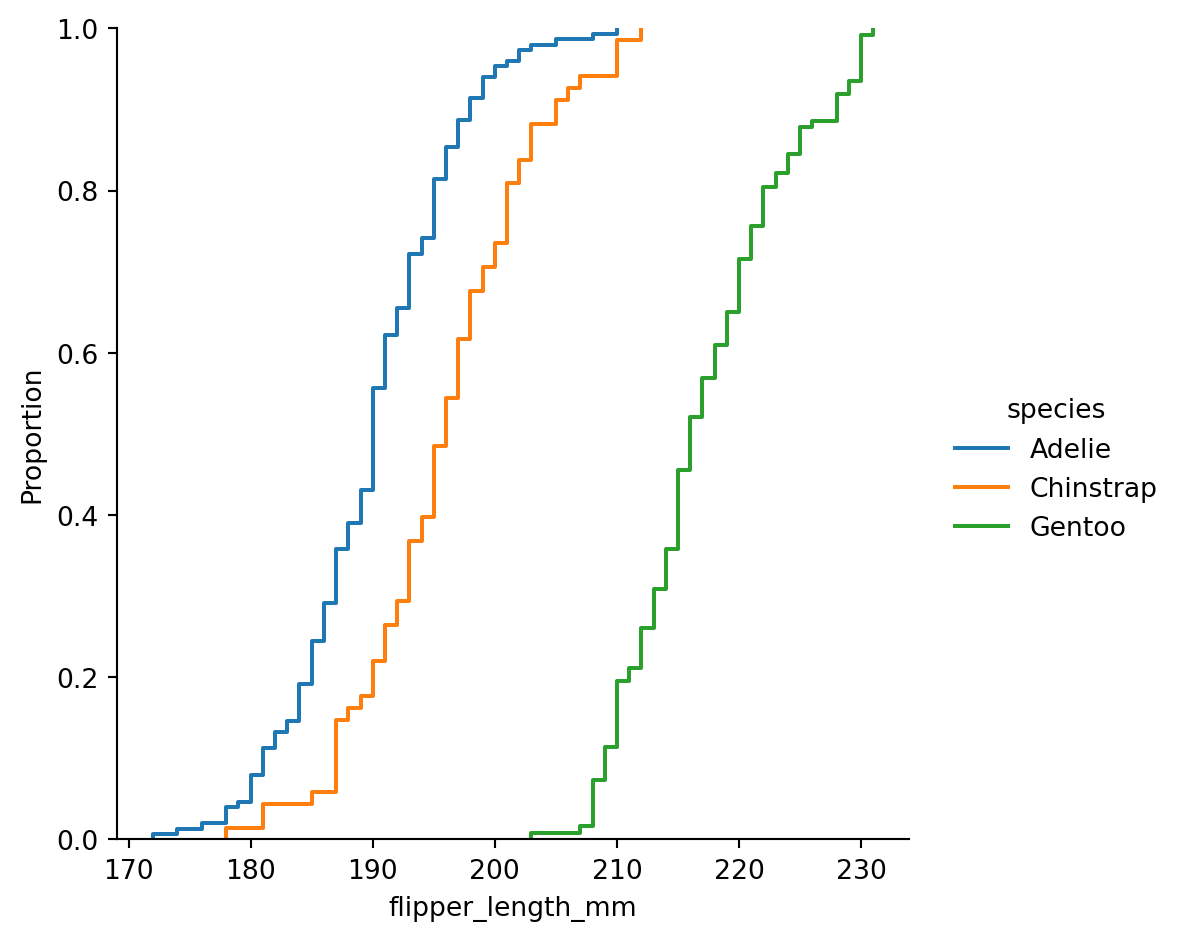
Empirical cumulative distribution - ecdfplot()
ECDF
This plot draws a monotonically-increasing curve through each datapoint such that the height of the curve reflects the proportion of observations with a smaller value.
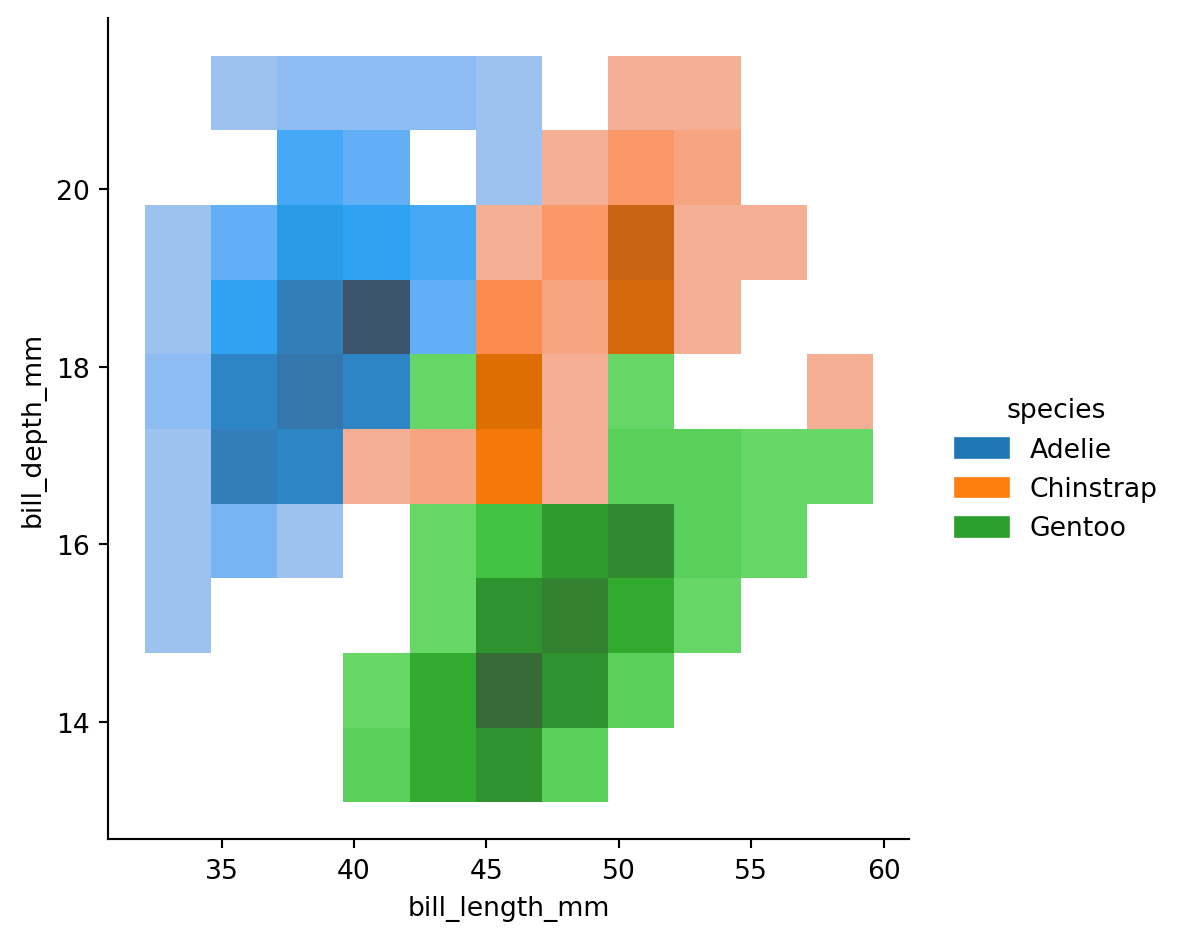
Visualizing bivariate distributions
Assigning a second variable to
y, however, will plot a bivariate distribution.Analogous to a
heatmap()
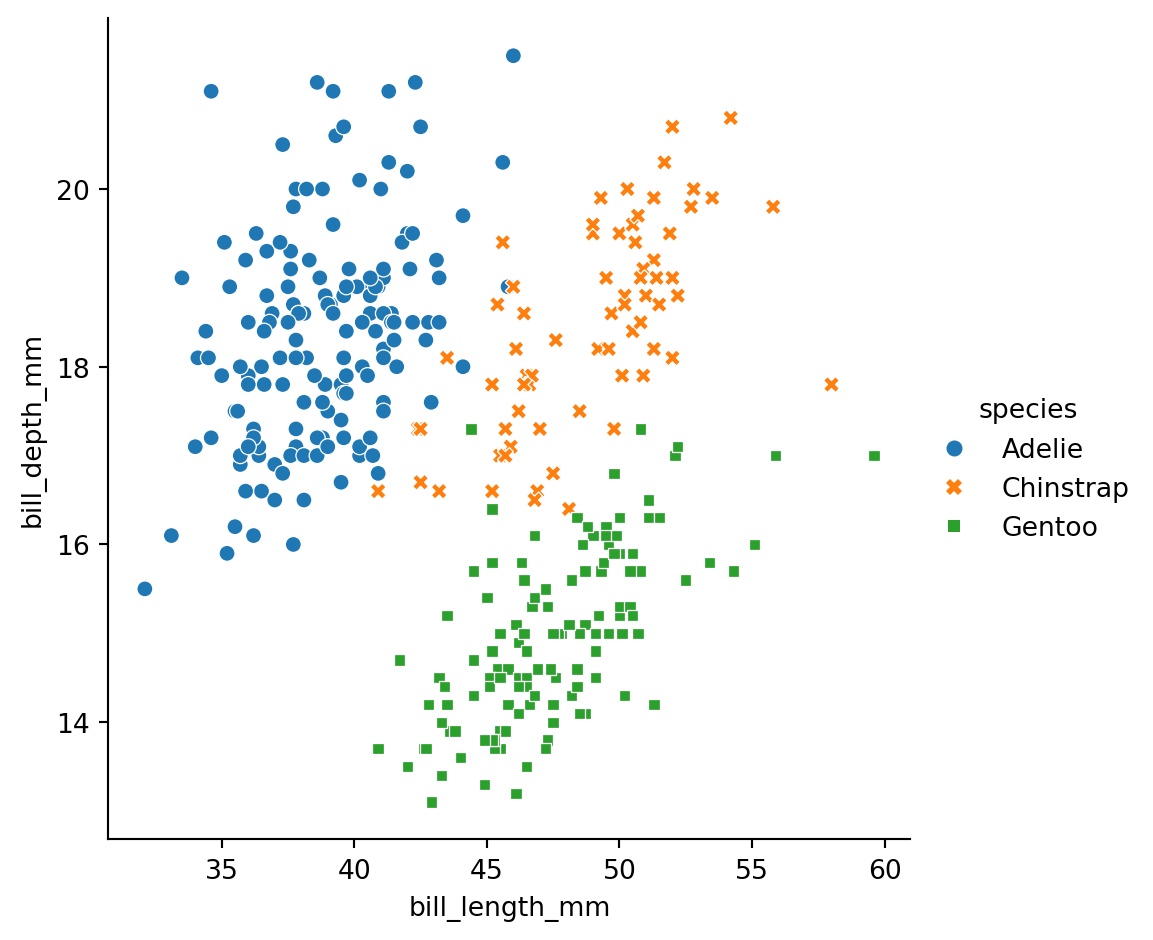
Scatter plots
Line plots
Note
The default behavior in seaborn is to aggregate the multiple measurements at each x value by plotting the mean and the 95% confidence interval around the mean.
Is this a good plot?
What is the connection between the three penguins species?
Categorical plots
Whilst for relational plots the main relationship is between two numerical variables, if one of the main variables is “categorical” (divided into discrete groups) it may be helpful to use a more specialized approach to visualization.
| Scatterplots | Distribution | Estimate |
|---|---|---|
stripplot() |
boxplot() |
pointplot() |
swarmplot() |
violinplot() |
barplot() |
| - | boxenplot() |
countplot() |
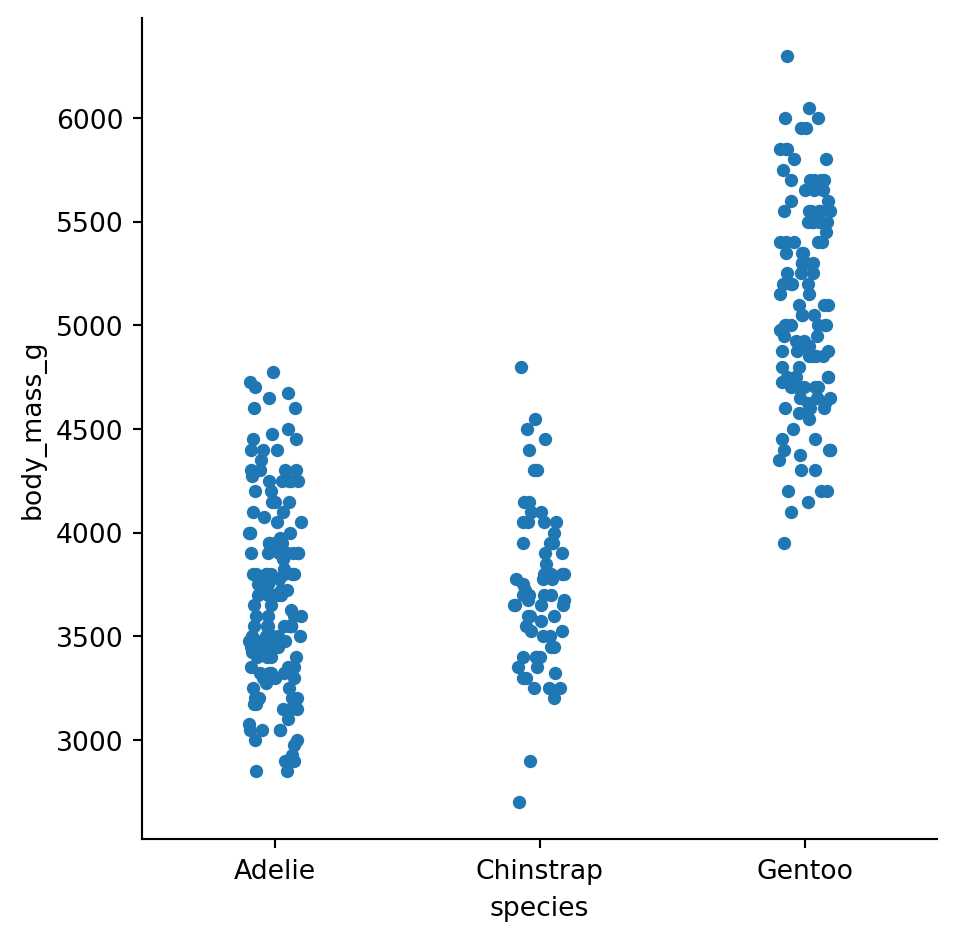
Categorical scatterplots - striplot()
Avoiding overplotting
Positions on the categorical axis receive a small amount of random jitter for better display of density.
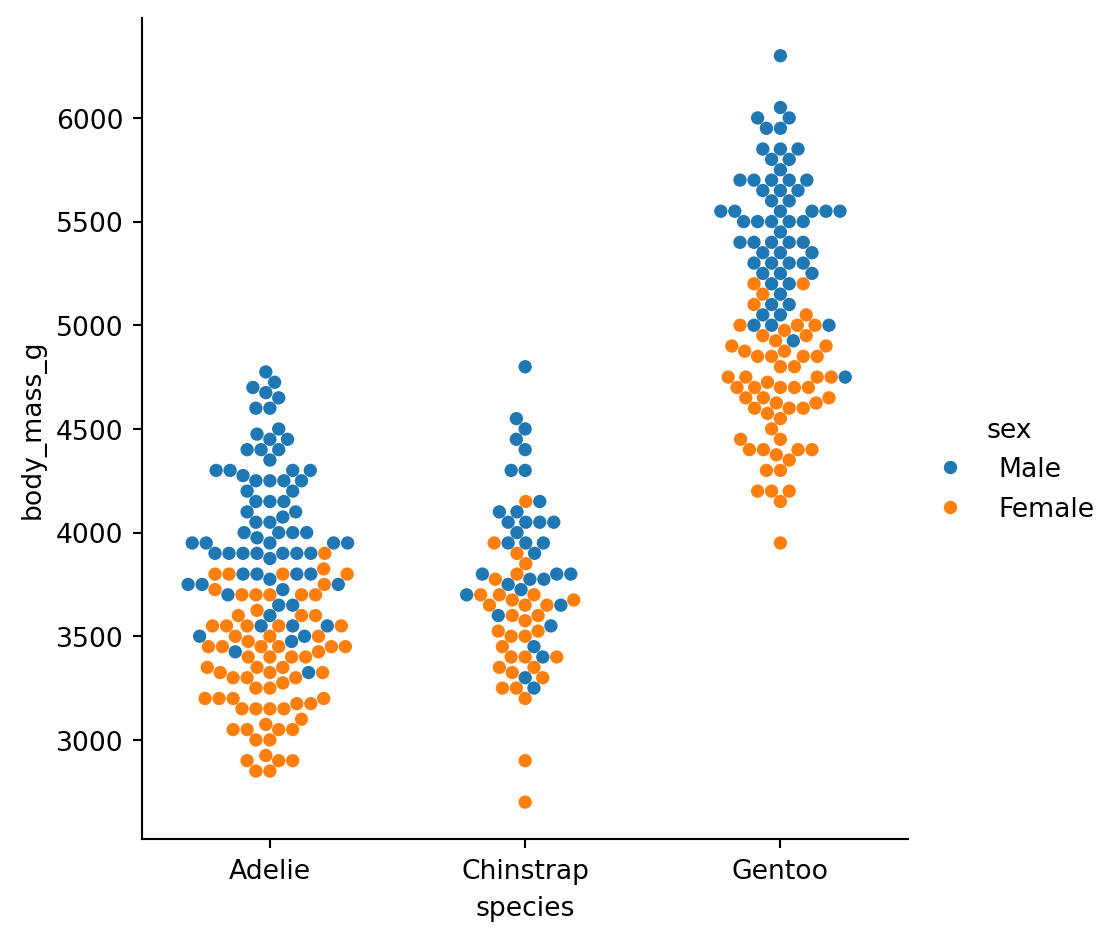
Categorical scatterplots swarmplot()
Beeswarm to show all points
It adjusts the points along the categorical axis using an algorithm that prevents them from overlapping.
Categorical distributions
Categorical scatter plots become limited as the dataset increases.
On those cases, distributions facilitate comparisons across the category levels.
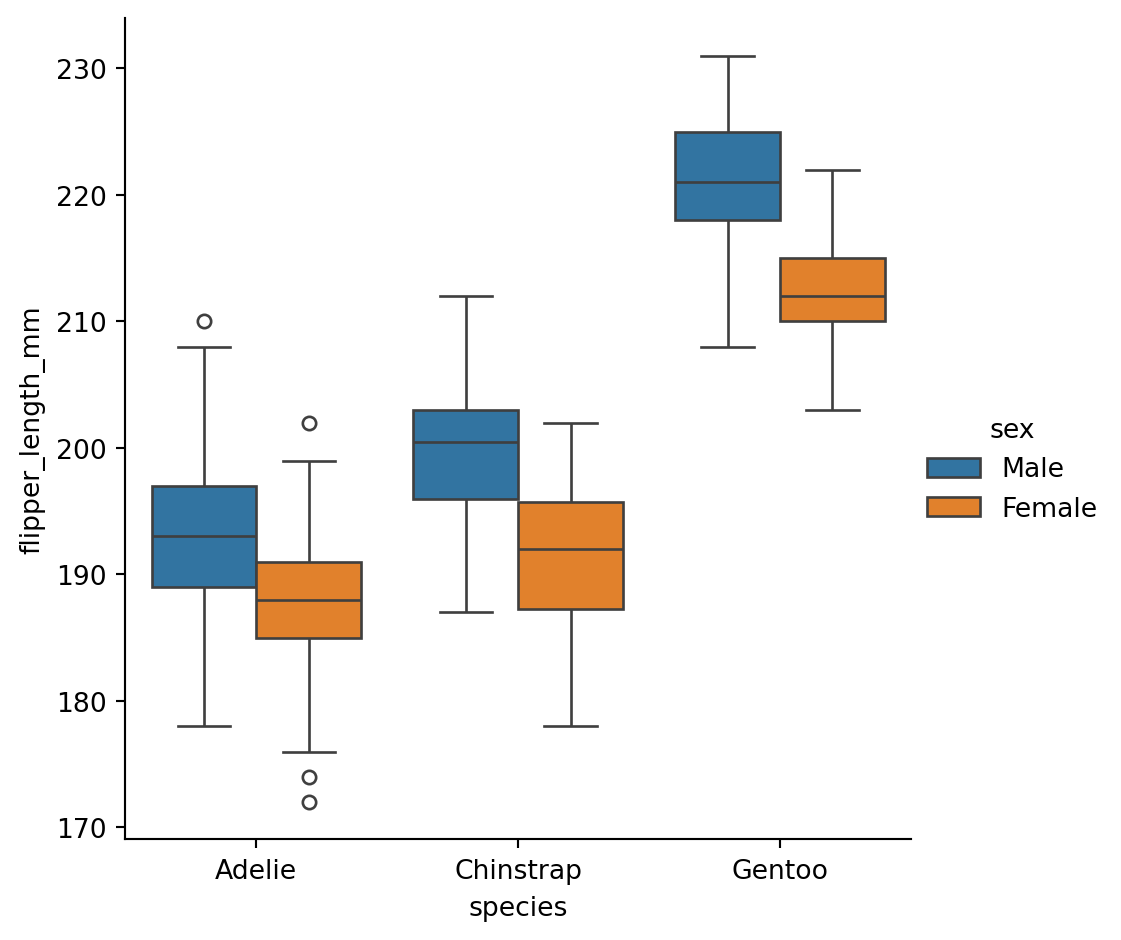
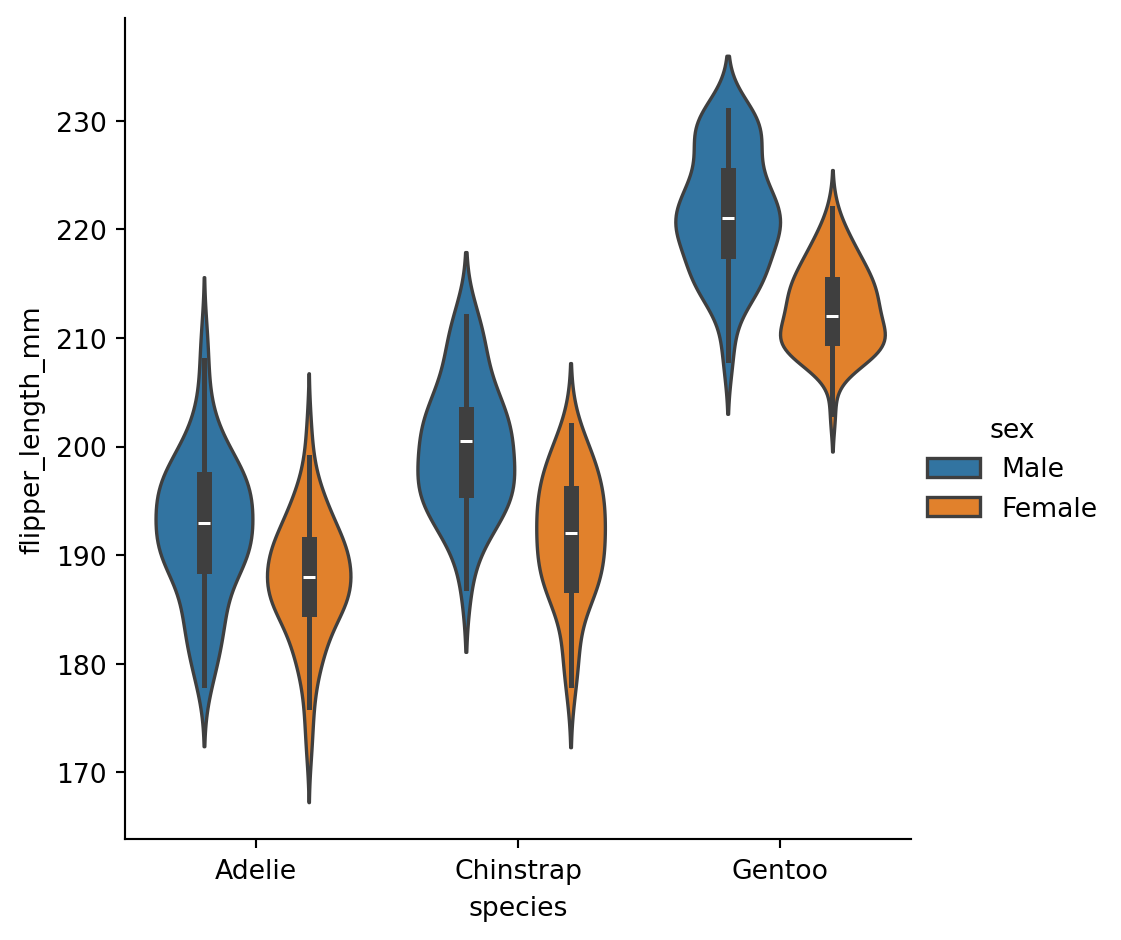
Boxplots
What’s in the box?
The three quartile values of the distribution along with extreme values, minimum and maximum data point.
Whiskers extend to points that lie within 1.5 IQRs of the lower and upper quartile. Observations that fall outside this range are displayed independently.
Boxplots in R
ggplot(penguins |> filter(!is.na(sex))) +
aes(x = species, y = flipper_length_mm, fill = fct_inorder(sex)) +
geom_boxplot(notch = TRUE)R uses the same standard settings for measures and whiskers settings (1.5 IQR) ending in a point.
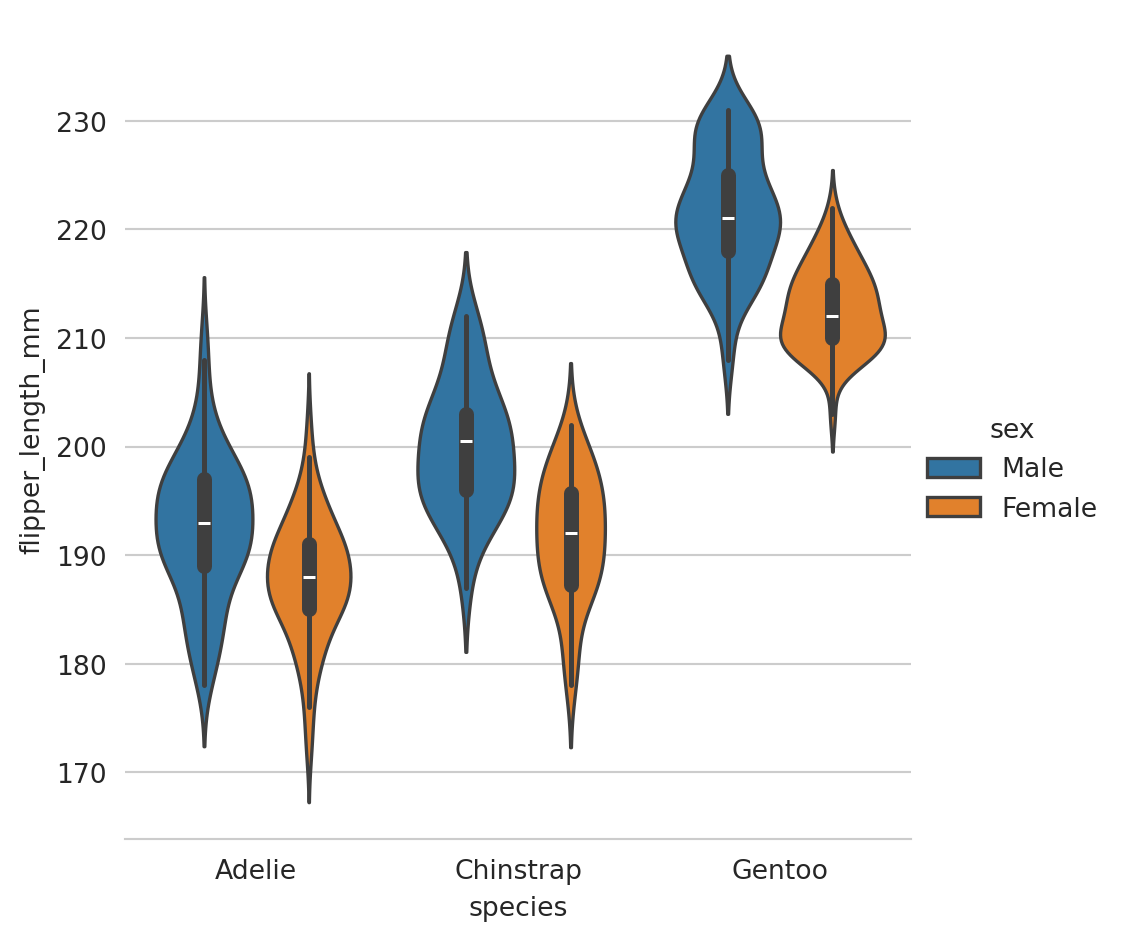
Violinplots
A violinplot() combines a boxplot with the kernel density estimation procedure.
Categorical estimates
In some cases, an estimate of the central tendency of the values would be better than only showing a distribution.
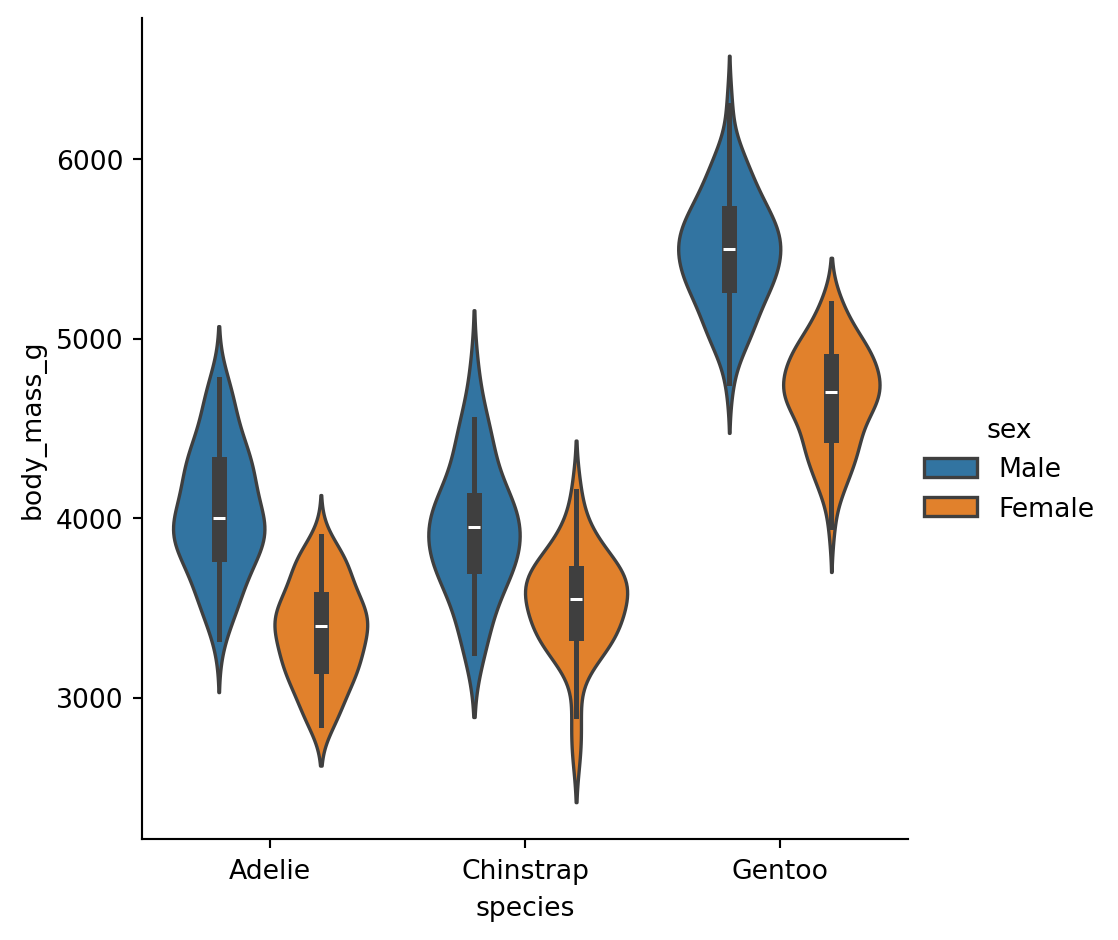
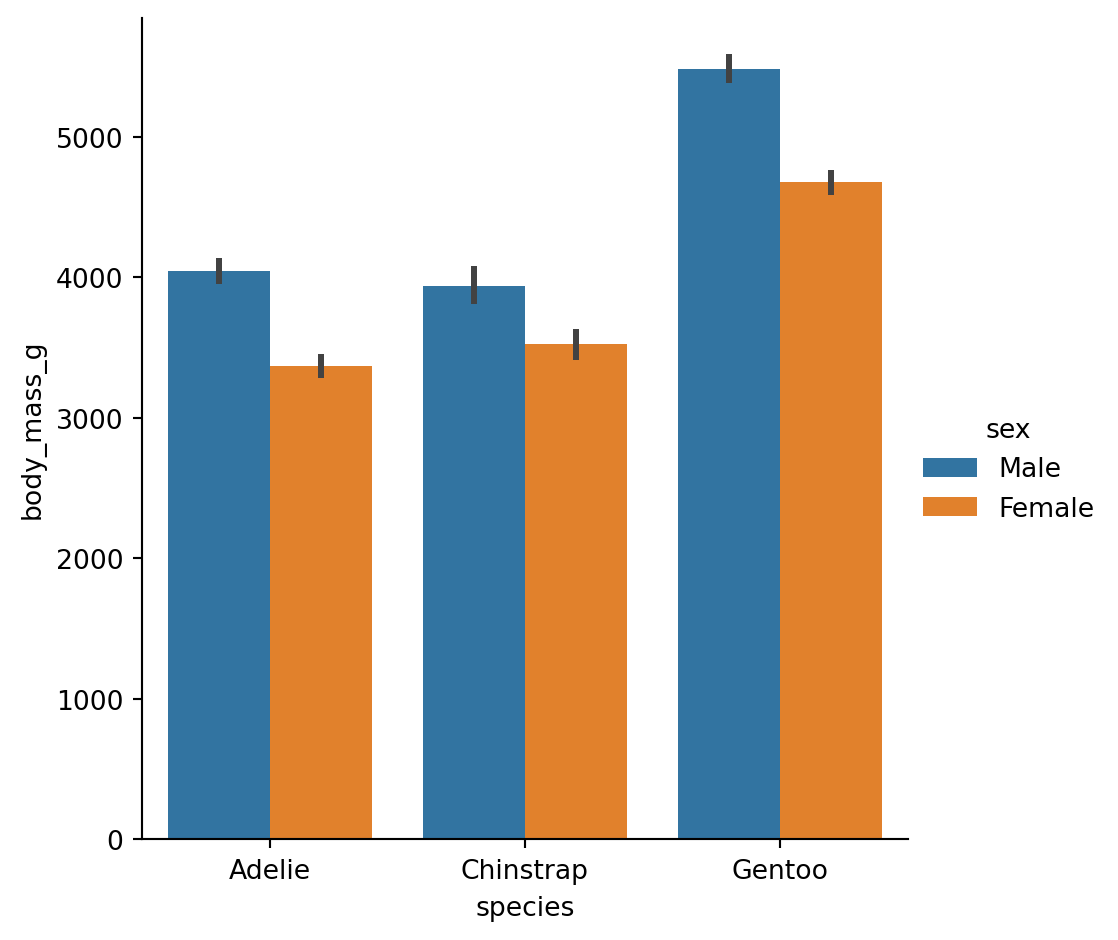
Bar plots
A familiar style of plot that accomplishes this goal is a bar plot.
The
barplot()function operates on a full dataset and applies a function to obtain the estimate.
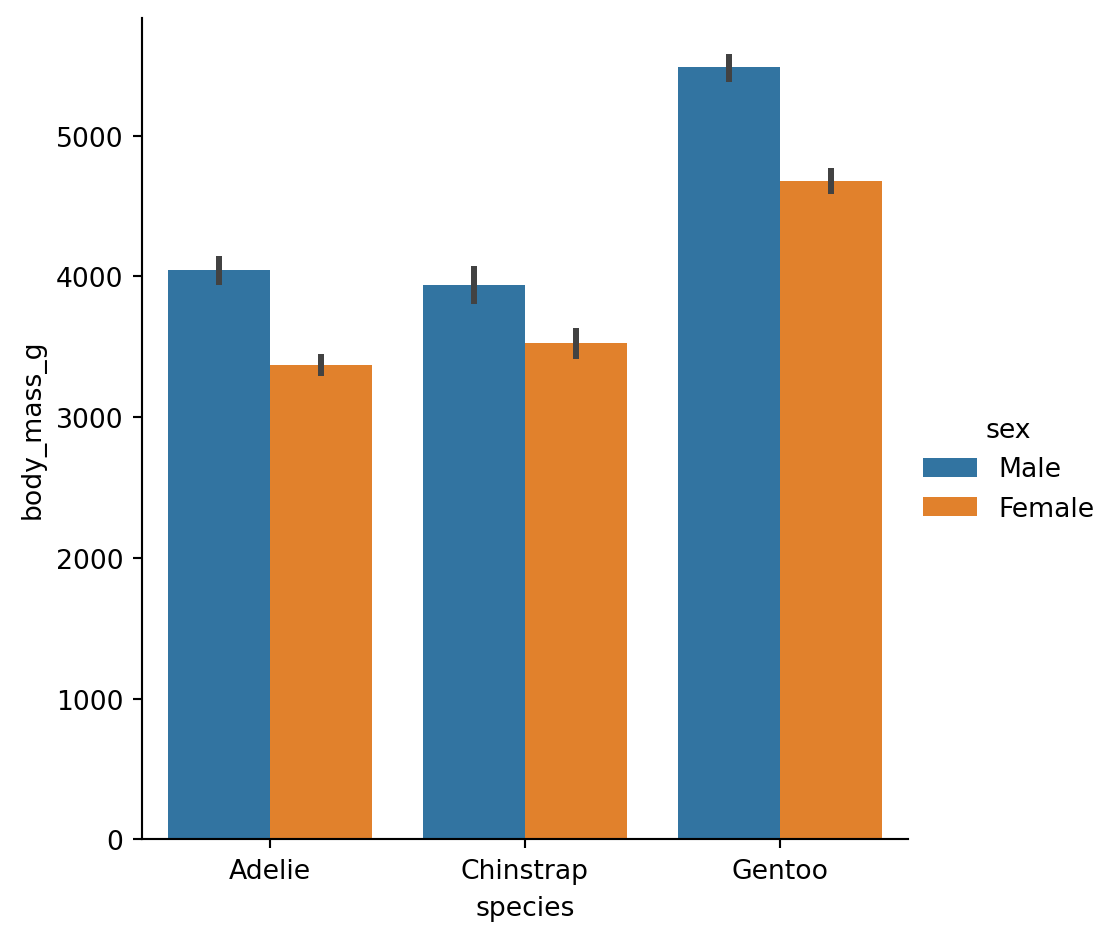
The danger of dynamite plots
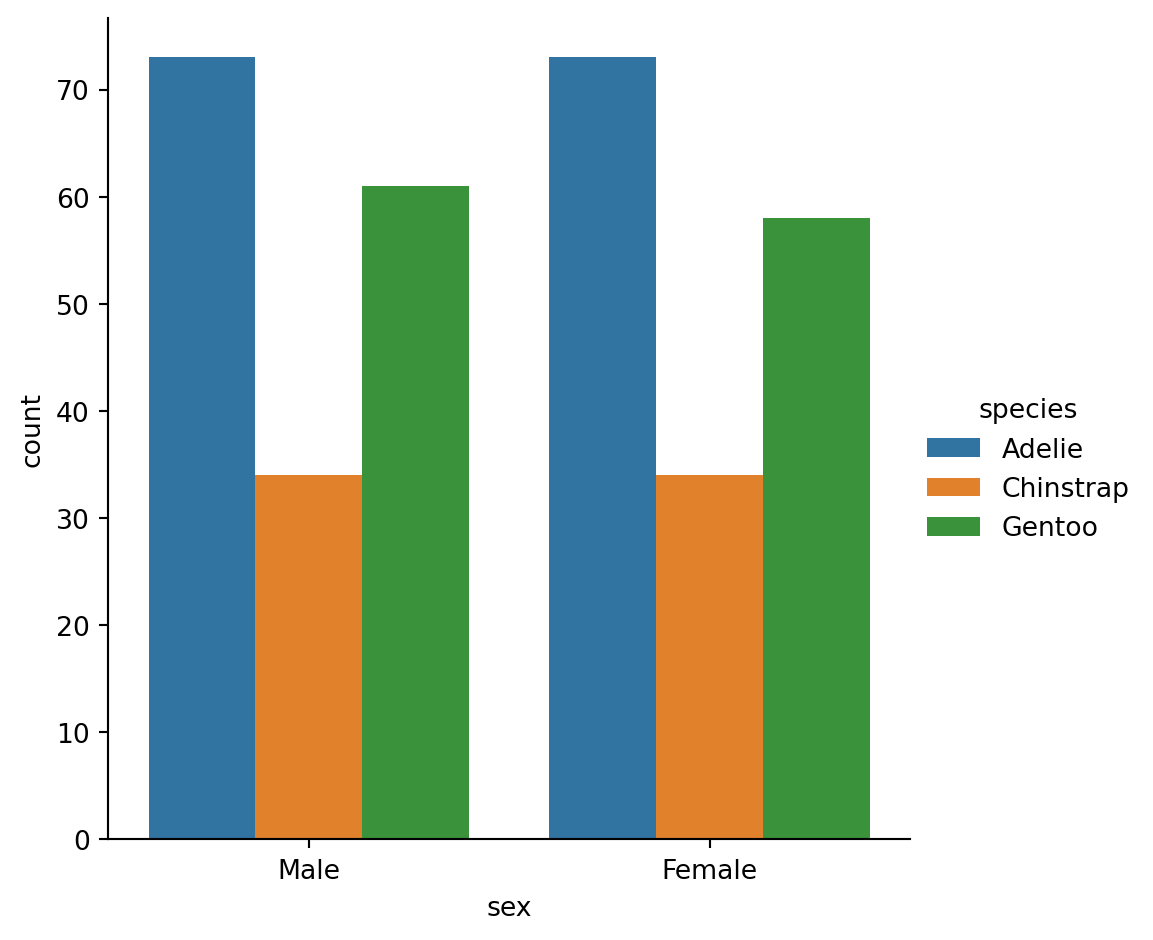
Count plots
- A special case for the bar plot is when you want to show the number of observations in each category rather than computing a statistic for a second variable.
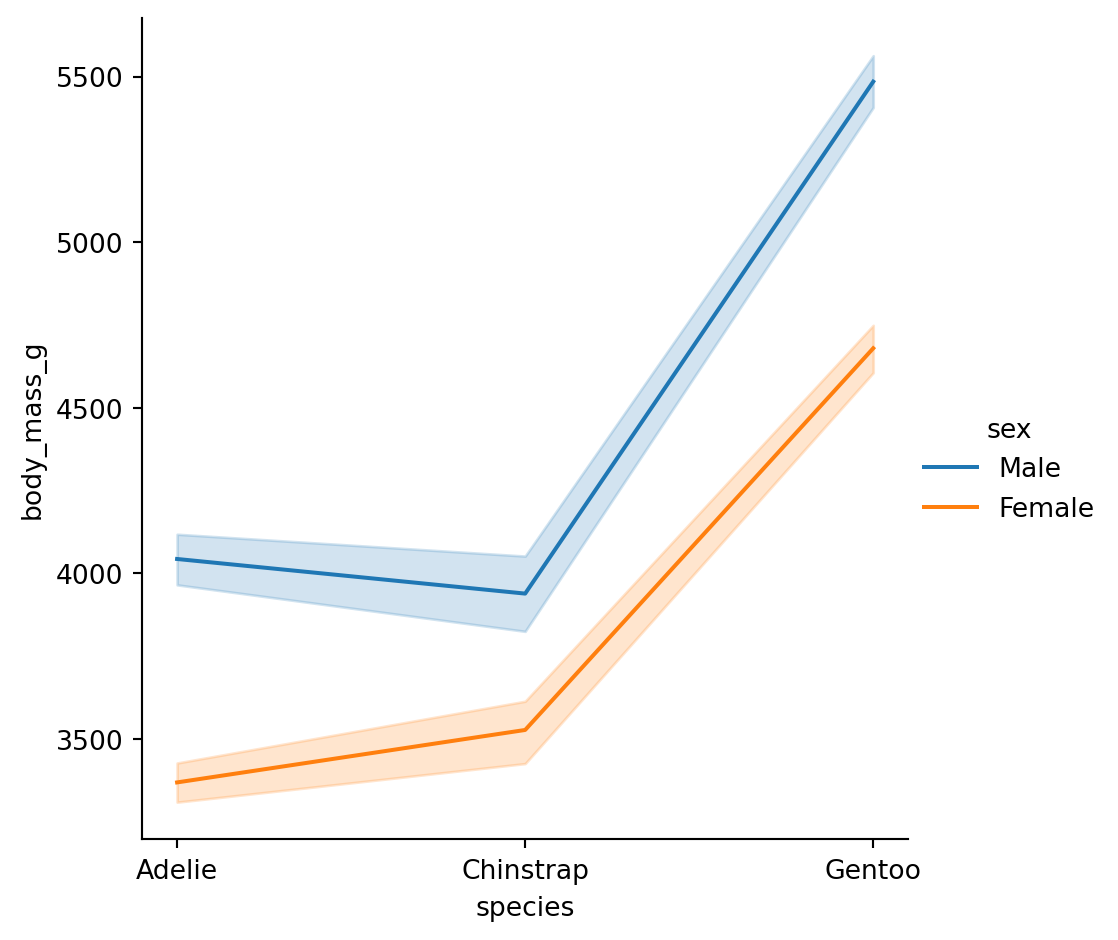
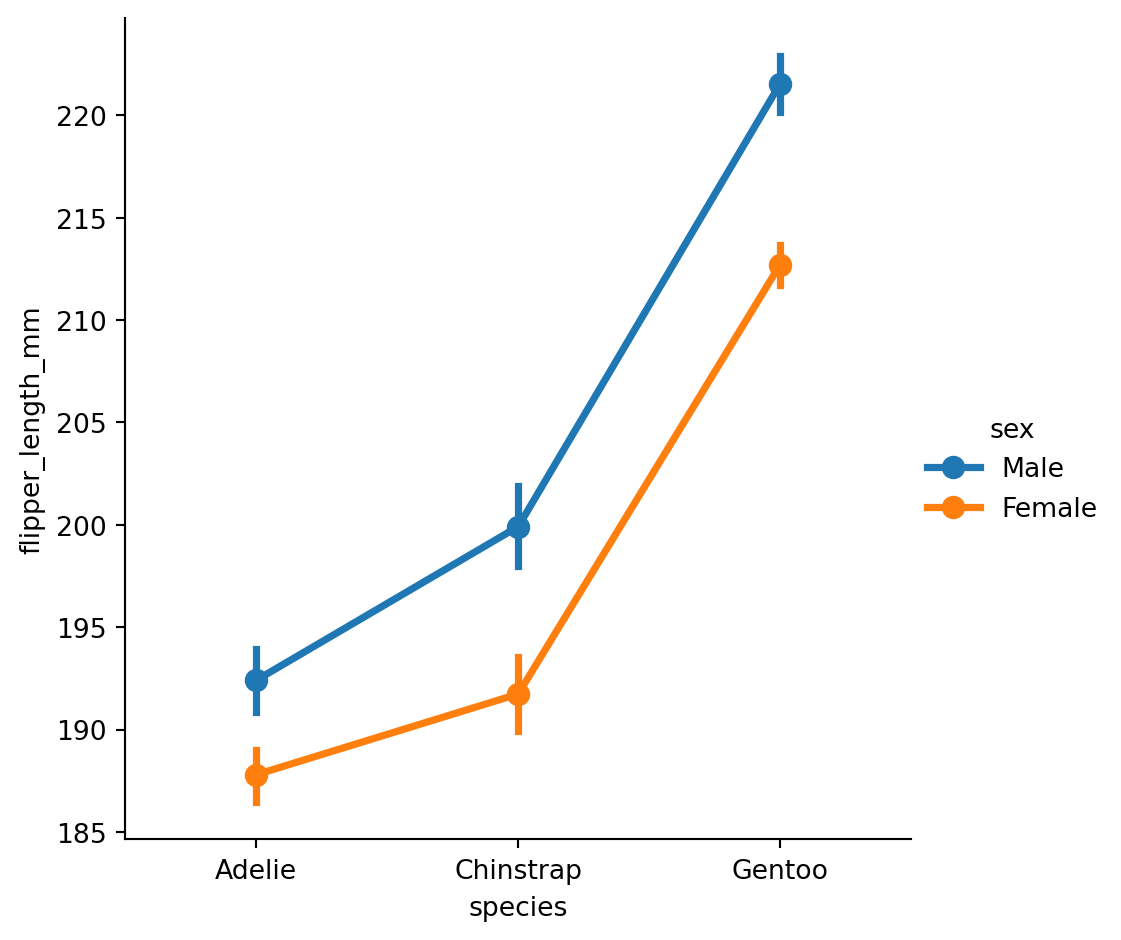
Point plots
An alternative style for visualizing the same information is offered by the
pointplot()function.It connects points from the same
huecategory which makes easy to see how the main relationship is changing as a function of thehuesemantic.
pointplot
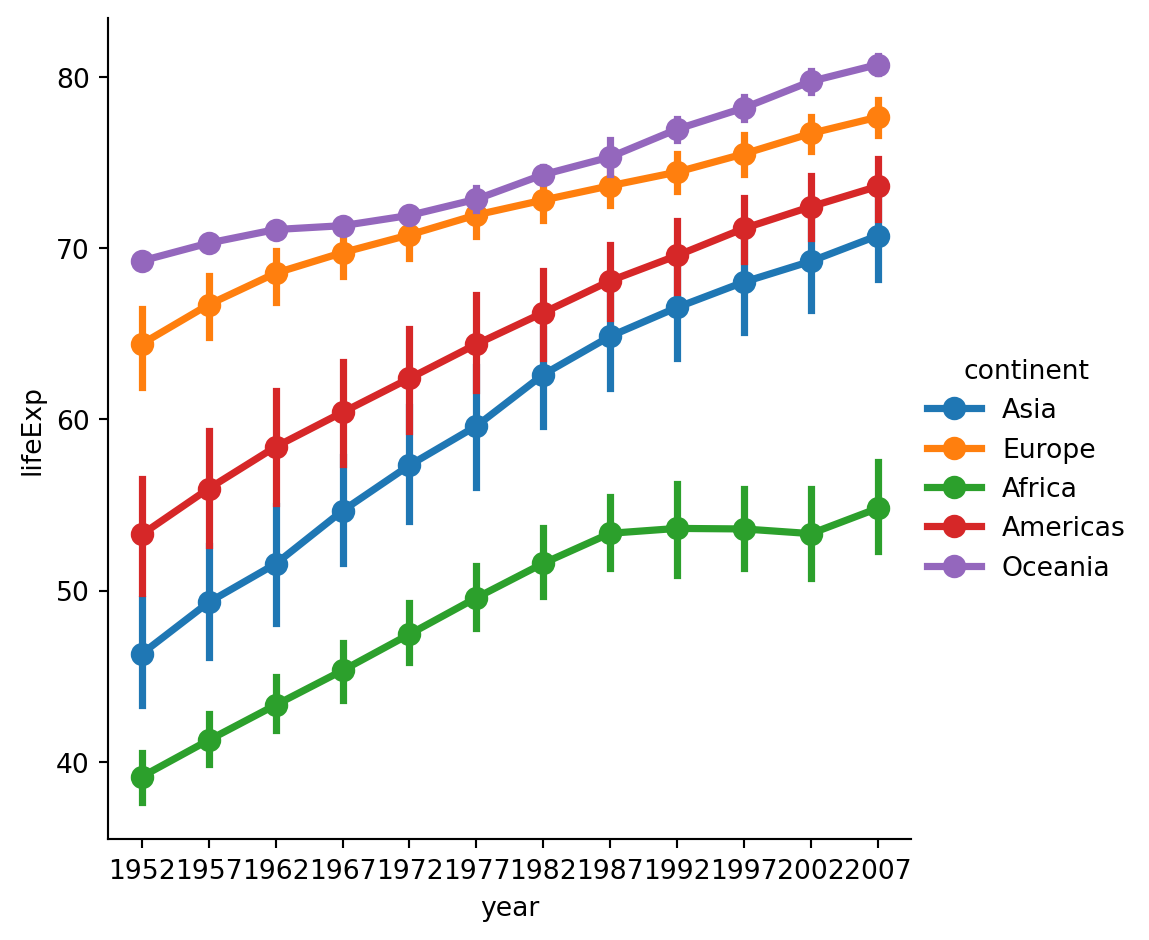
Direct labeling
From https://lost-stats.github.io/Presentation/Figures/line_graph_with_labels_at_the_beginning_or_end.html - not working because expects lines in columns
import numpy as np
fig, ax = plt.subplots()
sns.lineplot(ax=ax, data=gapminder[gapminder["country"] == "France"], x="year", y="lifeExp", hue = "continent", legend=None)
# sns.catplot(data = gapminder,
# x = "year",
# y = "lifeExp",
# hue = 'continent',
# kind = "point")
for line, name in zip(ax.lines, gapminder[gapminder["country"] == "France"].columns.tolist()):
y = line.get_ydata()[-1]
x = line.get_xdata()[-1]
if not np.isfinite(y):
y=next(reversed(line.get_ydata()[~line.get_ydata().mask]),float("nan"))
if not np.isfinite(y) or not np.isfinite(x):
continue
text = ax.annotate(name,
xy=(x, y),
xytext=(0, 0),
color=line.get_color(),
xycoords=(ax.get_xaxis_transform(),
ax.get_yaxis_transform()),
textcoords="offset points")
text_width = (text.get_window_extent(
fig.canvas.get_renderer()).transformed(ax.transData.inverted()).width)
if np.isfinite(text_width):
ax.set_xlim(ax.get_xlim()[0], text.xy[0] + text_width * 1.05)
plt.tight_layout()
plt.show()Figure vs axes level functions
Figure level
The figure-level functions can easily create figures with multiple subplots.
The kind-specific parameters don’t appear in the function signature or doc strings.
More complicated to set up fine adjustments.
Axes level
f, axs = plt.subplots(1, 2, figsize=(8, 4),
gridspec_kw=dict(width_ratios=[4, 3]))
sns.scatterplot(data=penguins,
x="flipper_length_mm",
y="bill_length_mm",
hue="species",
ax=axs[0])
sns.histplot(data=penguins,
x="species",
hue="species",
shrink=.8,
alpha=.8,
legend=False,
ax=axs[1])
f.tight_layout()
plt.show()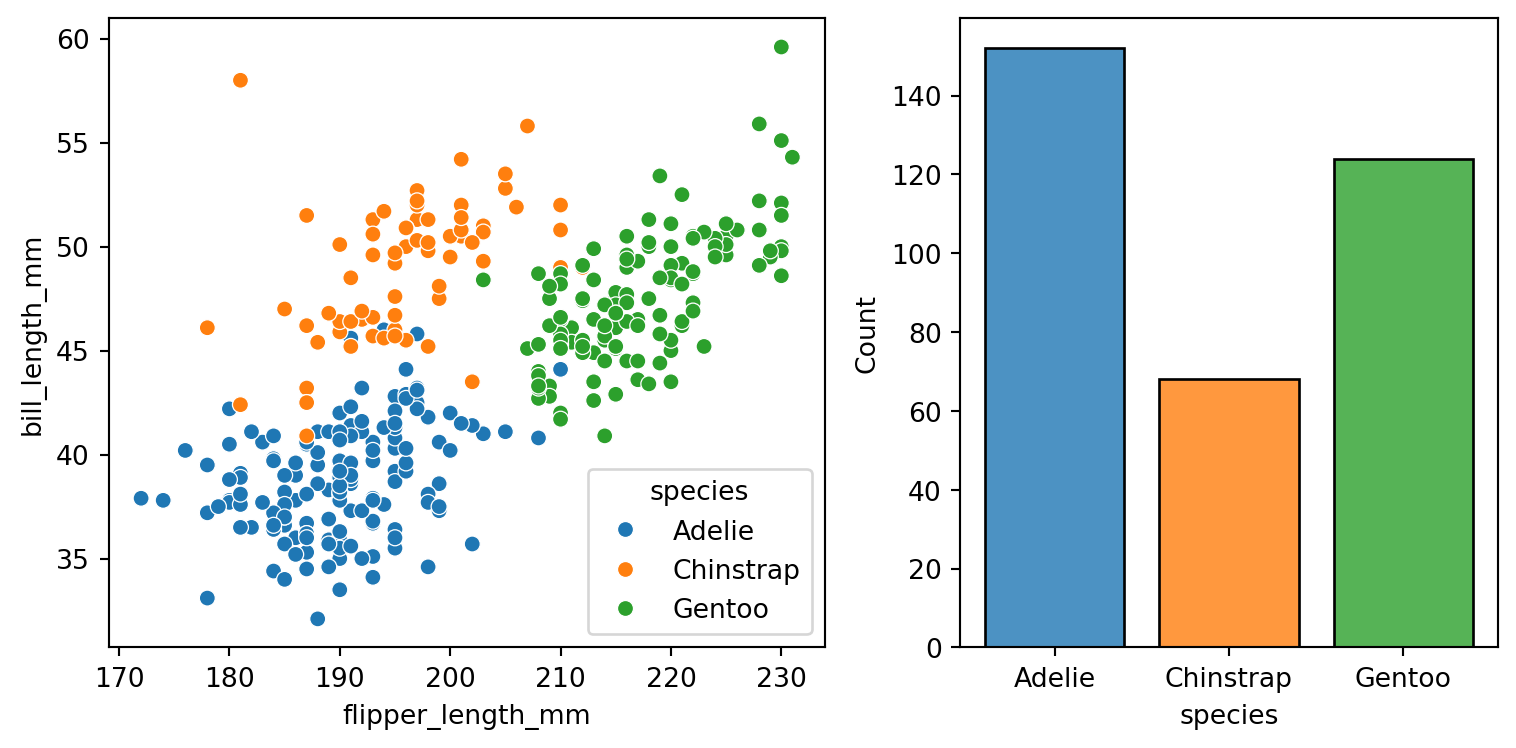
Axes-level functions don’t modify anything beyond the axes that they are drawn into.
Easier to compose into arbitrarily-complex matplotlib figures.
Multiple views on the data
There are two additional important functions that don’t fit cleanly into the classification scheme above.
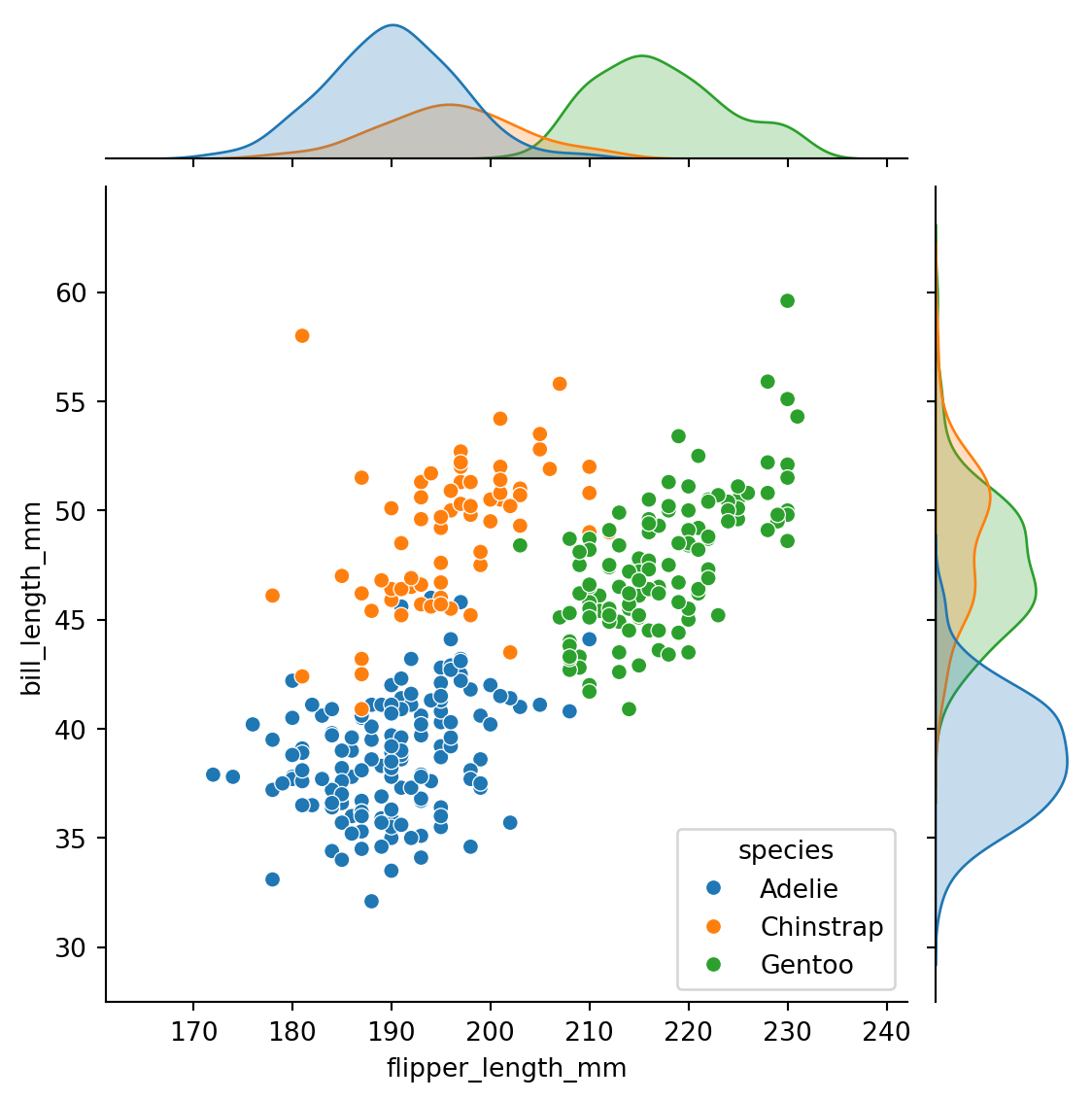
jointplot()
Plots the relationship or joint distribution of two variables while adding marginal axes that show the univariate distribution of each one separately:
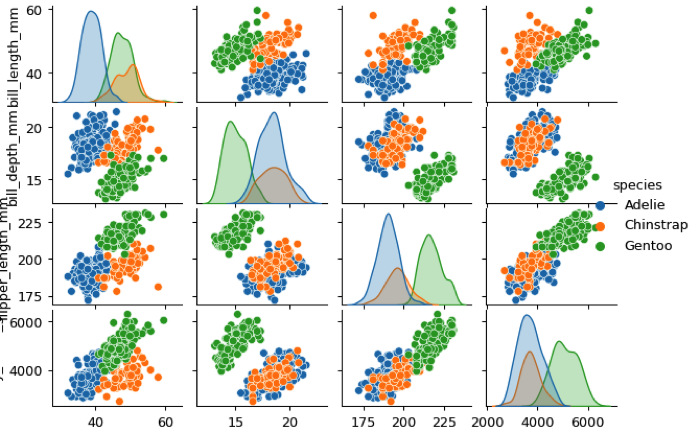
pairplot()
Figure aesthetics and context
Seaborn splits matplotlib parameters into two independent groups:
- Aesthetic
axes_style()andset_style(). - Context
plotting_context()andset_context().
Figure styles
rc parameter in the style functions
Parameter mappings to override the values in the preset Seaborn-style dictionaries.
Figure styles - temporary
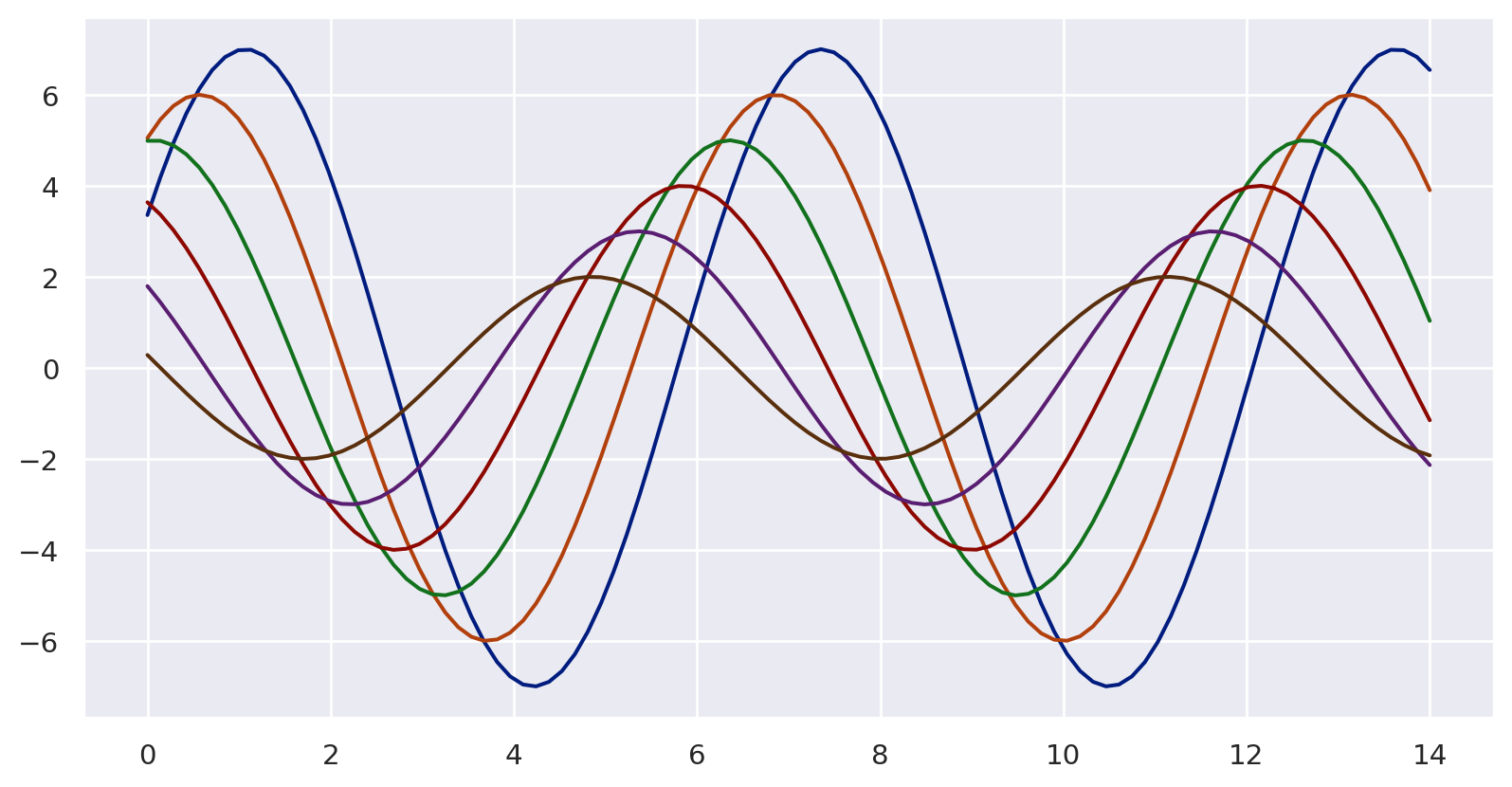
import numpy as np
f = plt.figure(figsize=(6, 6))
gs = f.add_gridspec(2, 2)
def sinplot(n=10, flip=1):
x = np.linspace(0, 14, 100)
for i in range(1, n + 1):
plt.plot(x, np.sin(x + i * .5) * (n + 2 - i) * flip)
with sns.axes_style("darkgrid"):
ax = f.add_subplot(gs[0, 0])
sinplot(6)
with sns.axes_style("white"):
ax = f.add_subplot(gs[0, 1])
sinplot(6)
with sns.axes_style("ticks"):
ax = f.add_subplot(gs[1, 0])
sinplot(6)
with sns.axes_style("whitegrid"):
ax = f.add_subplot(gs[1, 1])
sinplot(6)
f.tight_layout()
plt.show()
Controling spines
The white and ticks styles can benefit from removing the top and right axes spines:
Context plots
Control the scale of plot elements. The four preset contexts, in order of relative size, are paper, notebook, talk, and poster.
Context plots
Color palettes
- Color can reveal patterns in data if used effectively or hide patterns if used poorly.
seaborn.color_palette([palette], [n_colors], [desat])
Categorical color palettes
Best suited for distinguishing categorical data that does not have an inherent ordering.
The color palette should have colors as distinct from one another as possible.
Six default themes in Seaborn:
deep,muted,bright,pastel,dark, andcolorblind.
Sequential Color Palettes
- Appropriate for sequential data ranges from low to high values, or vice versa.
- Some suggest to use bright colors for low values and dark ones for high values; eventually it’s context-dependent.
Sequential palettes for heatmaps
Diverging Color Palettes
- Used for data that consists of a well-defined midpoint.
- An emphasis is placed on both high and low values.
Plotly

The plotly library is an interactive, open-source plotting library that supports over 40 unique chart types covering a wide range of statistical, financial, geographic, scientific, and 3-dimensional use-cases.
Built on top of the Plotly JavaScript library (plotly.js), plotly enables Python users to create beautiful interactive web-based visualizations that can be displayed in Jupyter notebooks, saved to standalone HTML files, or served as part of pure Python-built web applications using Dash.
Setup for plotly usage
Restaurant tips data
Food servers’ tips in restaurants may be influenced by many factors, including the nature of the restaurant, size of the party, and table locations in the restaurant.

Load the data
In one restaurant, a food server recorded the following data on all customers they served during an interval of two and a half months in early 1990.
| total_bill | tip | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 1.01 | Female | No | Sun | Dinner | 2 |
| 1 | 10.34 | 1.66 | Male | No | Sun | Dinner | 3 |
| 2 | 21.01 | 3.50 | Male | No | Sun | Dinner | 3 |
| 3 | 23.68 | 3.31 | Male | No | Sun | Dinner | 2 |
| 4 | 24.59 | 3.61 | Female | No | Sun | Dinner | 4 |
Distribution plots
Histograms
Plotly histograms will automatically bin numerical or date data.
Normalized histograms
The default mode is to represent the count of samples in each bin.
With the
histnormargument, it is also possible to represent the percentage or fraction of samples in each bin (histnorm='percent'or'probability').
Fine tuning a histogram
Combined histograms
Bivariate distributions
Scatter plots
Categorical scatter plots
Box plots
Violin plots
Bar plots
Point plots
Map plot
Using Python packages from R
reticulate

Comprehensive set of tools for interoperability between Python and R.
Calling Python from R in a variety of ways
Translation between R and Python objects
Flexible binding to different versions of Python including virtual environments and Conda environments.
Reticulate embeds a Python session within your R session, enabling seamless, high-performance interoperability.
Usage
By default
reticulatewill use the python version that is found on your PATH.You can use the
use_python()function to specify a different path to your python binary.As an alternative you can create a
condaenvironment containing your desired packages. (Recommended)In Quarto documents, Python code can be run side-by-side
Example in R
Before we stop
We learned to
- Use
matplotlibfor basic Python plots - Use Seaborn for fast, presentable plots
- Use plotly for data exploration
- Run Python inside R
Contributions
Izabela Ferreira da Silva (Original author)
Other materials